 |
PDBsum entry 1w0d

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1w0d
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
The high resolution structure of mycobacterium tuberculosis leub (rv2995c)
|
|
Structure:
|
 |
3-isopropylmalate dehydrogenase. Chain: a, b, c, d. Synonym: beta-ipm dehydrogenase, imdh, 3-ipm-dh. Engineered: yes. Other_details: rossman fold
|
|
Source:
|
 |
Mycobacterium tuberculosis. Organism_taxid: 83332. Strain: h37rv. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Biol. unit:
|
 |
 Dimer (from PDB file)
Dimer (from PDB file)
|
|
Resolution:
|
 |
|
1.65Å
|
R-factor:
|
0.213
|
R-free:
|
0.241
|
|
|
Authors:
|
 |
R.K.Singh,G.Kefala,R.Janowski,C.Mueller-Dieckmann,M.S.Weiss,Tb Structural Genomics Consortium (Tbsgc)
|
Key ref:

|
 |
R.K.Singh
et al.
(2005).
The high-resolution Structure of LeuB (Rv2995c) from Mycobacterium tuberculosis.
J Mol Biol,
346,
1.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
03-Jun-04
|
Release date:
|
14-Dec-04
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P9WKK9
(LEU3_MYCTU) -
3-isopropylmalate dehydrogenase from Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
336 a.a.
337 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.1.85
- 3-isopropylmalate dehydrogenase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Leucine Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(2R,3S)-3-isopropylmalate + NAD+ = 4-methyl-2-oxopentanoate + CO2 + NADH
|
 |
 |
 |
 |
 |
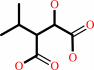 (2R,3S)-3-isopropylmalate
(2R,3S)-3-isopropylmalate
|
+
|
 NAD(+)
NAD(+)
|
=
|
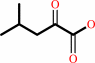 4-methyl-2-oxopentanoate
4-methyl-2-oxopentanoate
|
+
|
 CO2
CO2
|
+
|
 NADH
NADH
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
346:1
(2005)
|
|
PubMed id:
|
|
|
|

|
| |
|
The high-resolution Structure of LeuB (Rv2995c) from Mycobacterium tuberculosis.
|
|
R.K.Singh,
G.Kefala,
R.Janowski,
C.Mueller-Dieckmann,
J.P.von Kries,
M.S.Weiss.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The crystal structure of the enzyme 3-isopropylmalate dehydrogenase (IPMDH) from
Mycobacterium tuberculosis (LeuB, Mtb-IPMDH, Rv2995c) without substrate or
co-factor was determined at 1.65 A resolution, which is the highest resolution
reported for an IPMDH to date. The crystals contain two functional dimers in the
asymmetric unit in an arrangement close to a tetramer of D2 symmetry. Despite
the absence of a substrate or inhibitor bound to the protein, the structure of
the monomer resembles the previously observed closed form of the enzyme more
closely than the open form. A comparison with the substrate complex of IPMDH
from Thiobacillus ferrooxidans and the co-factor complex of the Thermus
thermophilus enzyme revealed a close relationship of the active-site
architecture between the various bacterial enzymes. The inhibitor O-isobutenyl
oxalylhydroxamate was found to bind to the active site of IPMDH in a mode
similar to the substrate isopropylmalate.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. The three-dimensional structure of Mtb-IPMDH. (a)
C^a representation of the four subunits of Mtb-IPMDH found in
the asymmetric unit. Each subunit is represented by a different
colour (A, red; B, green; C, yellow; D, cyan). The six local
dyads and the approximate local dyads are indicated. The
resemblance of the arrangement to a D[2] tetramer is clearly
discernible. (b) Ribbon representation of the structure of the
homodimer of Mtb-IPMDH. The two domains in one subunit are
identified by different colours, domain 1 is displayed in red,
domain 2 in blue. The two different shades of red in domain 1
denote the two sub-domains (see the text). (c) Ribbon
representation of the structure of one subunit of Mtb-IPMDH. The
secondary structure elements are coloured with a-helices in red
and b-strands in yellow. The N and C termini are labelled. The
1.0 kb gene fragment of Rv2995c was amplified by PCR from M.
tuberculosis H37Rv genomic DNA with the following
oligonucleotides (obtained from MWG Biotech):
5'-AAAATCATGAGCAAACTCGCGATCATTGCCGGTGACGGGATCGGGC-3' and
5'-AAAACTCGAGTTAGAGCGCGGCGGCAATCCGTTCGCCGACA-3' as forward and
reverse primers, respectively. In the forward primer, the
additional underlined bases were introduced to get the gene
in-frame for transcription. Therefore, the gene product obtained
contains an extra serine residue at position 2 (codon AGC). The
amplified fragment containing 5'-BspHI and 3'-XhoI restriction
sites (shown in bold in the primer sequence) was cloned into
PCR-Blunt TOPO vector (Invitrogen). The construct was digested
with BspHI and XhoI, and the insert fragment was subcloned into
the pETM-11 expression vector digested with the restriction
enzymes NcoI and XhoI. Both TOPO and the final pETM-11
constructs were sequenced to confirm the cloning of the leuB
gene sequence in-frame. Expression of the 35 kDa protein was
carried out in E. coli BL21 Star (DE3) pRARE cells. These cells
were prepared by transforming BL21 Star (DE3) cells (Invitrogen)
with the pRARE plasmid isolated from the Rosetta strain
(Novagen). Cells from an overnight pre-culture were grown in LB
broth medium containing kanamycin (30 µg/ml) and
chloramphenicol (10 µg/ml) at 310 K and 200 rpm. The
culture was induced with 0.2 mM isopropyl
b-D-thiogalactopyranoside (IPTG) at an A[600] of approximately
0.6 at 293 K. Following induction, the culture was incubated for
about 15 hours at 293 K and 200 rpm and then harvested. The
cells were stored at 193 K until further processing. One gram of
cell pellet was lysed by sonication in 30 ml of buffer A (50 mM
Tris (pH 8.0), 200 mM NaCl, 10 mM imidazole, 5% (v/v) glycerol,
2 mM b-mercaptoethanol (b-ME)) for 20 minutes in 25 second
pulses at 4 °C. The cell debris was pelleted by
centrifugation for 30 minutes at 4 °C and 20,000 rpm. The
crude lysate was filtered through a 0.2 µm pore size
membrane and loaded onto a 5 ml Hi-Trap Chelating HP column
(Amersham Biosciences) charged and equilibrated with Ni2+ and
buffer A, respectively. In order to remove unbound proteins, the
column was first washed with 25 ml of buffer A, then with 25 ml
of buffer B (50 mM Tris (pH 8.0), 600 mM NaCl, 10 mM imidazole,
5% (v/v) glycerol, 2 mM b-ME) and then with 25 ml of buffer C
(50 mM Tris (pH 8.0), 200 mM NaCl, 50 mM imidazole, 5% (v/v)
glycerol, 2 mM b-ME). The protein was eluted by running a linear
gradient from 50 mM to 800 mM imidazole (in buffer C) in a total
volume of 50 ml. The peak fractions were pooled and dialysed
against buffer D (50 mM Tris (pH 8.0), 200 mM NaCl, 5% (v/v)
glycerol, 2 mM b-ME). During dialysis overnight at 277 K, the
His tag was cleaved off by adding recombinant tobacco etch virus
(TEV) protease. The cleaved and dialysed protein mixture was
passed through a Ni-NTA column pre-equilibrated with buffer D.
The protein in the flow-through was subsequently purified by
gel-filtration (Superdex 200 16/60, Amersham Biosciences) using
buffer D for both equilibration and elution. The protein eluted
with an apparent molecular mass of approximately 70 kDa, which
indicated the presence of a homodimer in solution. The peak
fractions analysed by SDS-PAGE were pooled, and concentrated to
10 mg/ml. The purity of the protein was estimated by SDS-PAGE
and by dynamic light-scattering. The initial crystallization
screening was done in Greiner 96-well plates using the
sitting-drop, vapour-diffusion method (1 µl of protein and
1 µl of Hampton crystal screens 1 and 2). A few needles
were observed in condition #32 of screen #2 (1.6 M
(NH[4])[2]SO[4], 0.1 M Hepes (pH 7.5), 0.1 M NaCl) at 293 K.
This condition was optimized to 2.0 M (NH[4])[2]SO[4], 0.1 M
Hepes (pH 8.0), 0.1 M NaCl. The crystallization experiments
using the optimized condition as reservoir solution were
performed in 24-well plates using the hanging-drop method with
1.5 µl of protein and 1 µl of reservoir solution at
293 K. Crystals were typically obtained within 48 hours and grew
up to 1.0 mm in size. The crystals belong to space group
P2[1]2[1]2[1] with unit cell dimensions of a=78.47 Å,
b=97.06 Å and c=181.91 Å, and have four molecules in
the asymmetric unit. They diffract X-rays to about 1.5 Å
resolution. Crystals were treated with cryoprotectant (22% (v/v)
glycerol in reservoir solution) and diffraction data were
collected at 100 K on the EMBL wiggler beamline BW7B (DESY,
Hamburg, Germany) using a MAR345 mm imaging plate detector. Two
sweeps of data were collected: a low-resolution sweep (180°)
extending to 3.0 Å resolution and a high-resolution sweep
(180°) extending to 1.65 Å resolution. The data were
indexed and integrated using DENZO23 and scaled using
SCALEPACK.23 The redundancy-independent merging R-factor
R[r.i.m.] as well as the precision-indicating merging R-factor
R[p.i.m.]24 were calculated using the program RMERGE (available
from http://www.embl-hamburg.de/ msweiss/projects/msw_qual.html
or from M.S.W. upon request). The relevant data processing
parameters are given in Table 2. Intensities were converted to
|
 |
Figure 4.
Figure 4. (a) A representation of the proposed IPM and
NAD^+ binding sites in Mtb-IPMDH. (b) A representation of the
proposed O-isobutenyl oxalylhydroxamate (O-ibOHA) binding site.
All amino acid residues contacting either of the three
substances at a distance of less than 4.5 Å are shown. The
program GOLD was used for the docking experiments.22 The
starting coordinates for IPM and the inhibitor O-ibOHA were
generated by superimposing the IPM complex of IPMDH of T.
ferrooxidans (PDB entry 1A0510) onto the AB dimer of Mtb-IPMDH.
The coordinates of the C3 atom of IPM were defined as the centre
of the search sphere. The search was then carried out within a
radius of 10 Å, using the default settings of GOLD,
including cavity detection and a limit of 50 different dockings.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2005,
346,
1-0)
copyright 2005.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
K.Manikandan,
A.Geerlof,
A.V.Zozulya,
D.I.Svergun,
and
M.S.Weiss
(2011).
Structural studies on the enzyme complex isopropylmalate isomerase (LeuCD) from Mycobacterium tuberculosis.
|
| |
Proteins,
79,
35-49.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Ã.‰.Gráczer,
A.Merli,
R.K.Singh,
M.Karuppasamy,
P.Závodszky,
M.S.Weiss,
and
M.Vas
(2011).
Atomic level description of the domain closure in a dimeric enzyme: thermus thermophilus 3-isopropylmalate dehydrogenase.
|
| |
Mol Biosyst,
7,
1646-1659.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
I.Hajdú,
A.Szilágyi,
J.Kardos,
and
P.Závodszky
(2009).
A link between hinge-bending domain motions and the temperature dependence of catalysis in 3-isopropylmalate dehydrogenase.
|
| |
Biophys J,
96,
5003-5012.
|
 |
|
|
|
|
 |
M.Karuppasamy,
A.Geerlof,
L.Schuldt,
C.Mueller-Dieckmann,
and
M.S.Weiss
(2009).
Cloning, expression, purification, crystallization and preliminary X-ray diffraction analysis of the small subunit of isopropylmalate isomerase (Rv2987c) from Mycobacterium tuberculosis.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
65,
136-139.
|
 |
|
|
|
|
 |
C.Mueller-Dieckmann,
S.Panjikar,
A.Schmidt,
S.Mueller,
J.Kuper,
A.Geerlof,
M.Wilmanns,
R.K.Singh,
P.A.Tucker,
and
M.S.Weiss
(2007).
On the routine use of soft X-rays in macromolecular crystallography. Part IV. Efficient determination of anomalous substructures in biomacromolecules using longer X-ray wavelengths.
|
| |
Acta Crystallogr D Biol Crystallogr,
63,
366-380.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.A.McCourt,
and
R.G.Duggleby
(2006).
Acetohydroxyacid synthase and its role in the biosynthetic pathway for branched-chain amino acids.
|
| |
Amino Acids,
31,
173-210.
|
 |
|
|
|
|
 |
V.L.Arcus,
J.S.Lott,
J.M.Johnston,
and
E.N.Baker
(2006).
The potential impact of structural genomics on tuberculosis drug discovery.
|
| |
Drug Discov Today,
11,
28-34.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

