 |
PDBsum entry 1qqj

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.7.1.2
- fumarylacetoacetase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
4-fumarylacetoacetate + H2O = acetoacetate + fumarate + H+
|
 |
 |
 |
 |
 |
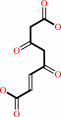 4-fumarylacetoacetate
4-fumarylacetoacetate
|
+
|
H2O
|
=
|
acetoacetate
Bound ligand (Het Group name = )
matches with 57.14% similarity
|
+
|
 fumarate
fumarate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Structure
7:1023-1033
(1999)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure and mechanism of a carbon-carbon bond hydrolase.
|
|
D.E.Timm,
H.A.Mueller,
P.Bhanumoorthy,
J.M.Harp,
G.J.Bunick.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
BACKGROUND: Fumarylacetoacetate hydrolase (FAH) catalyzes the final step of
tyrosine and phenylalanine catabolism, the hydrolytic cleavage of a
carbon-carbon bond in fumarylacetoacetate, to yield fumarate and acetoacetate.
FAH has no known sequence homologs and functions by an unknown mechanism.
Carbon-carbon hydrolysis reactions are essential for the human metabolism of
aromatic amino acids. FAH deficiency causes the fatal metabolic disease
hereditary tyrosinemia type I. Carbon-carbon bond hydrolysis is also important
in the microbial metabolism of aromatic compounds as part of the global carbon
cycle. RESULTS: The FAH crystal structure has been determined by rapid,
automated analysis of multiwavelength anomalous diffraction data. The FAH
polypeptide folds into a 120-residue N-terminal domain and a 300-residue
C-terminal domain. The C-terminal domain defines an unusual beta-strand topology
and a novel 'mixed beta-sandwich roll' structure. The structure of FAH complexed
with its physiological products was also determined. This structure reveals
fumarate binding near the entrance to the active site and acetoacetate binding
to an octahedrally coordinated calcium ion located in close proximity to a
Glu-His dyad. CONCLUSIONS: FAH represents the first structure of a hydrolase
that acts specifically on carbon-carbon bonds. FAH also defines a new class of
metalloenzymes characterized by a unique alpha/beta fold. A mechanism involving
a Glu-His-water catalytic triad is suggested based on structural observations,
sequence conservation and mutational analysis. The histidine imidazole group is
proposed to function as a general base. The Ca(2+) is proposed to function in
binding substrate, activating the nucleophile and stabilizing a carbanion
leaving group. An oxyanion hole formed from sidechains is proposed to stabilize
a tetrahedral alkoxide transition state. The proton transferred to the carbanion
leaving group is proposed to originate from a lysine sidechain. The results also
reveal the molecular basis for mutations causing the hereditary tyrosinemia type
1.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |
Figure 2.
Figure 2. FAH structure, topology and HT1-associated
mutations. (a) A stereo ribbon diagram illustrating the FAH
subunit structure and position of point mutations causing
hereditary tyrosinemia type I is shown. The N-terminal domain is
located at the bottom of the figure. The mixed b-sandwich roll
structure is centrally located in the figure. Helices are
colored red; b strands are colored in shades of blue
corresponding to the b sheet they form; the positions of point
mutations are represented by green spheres; a calcium ion is
colored yellow; acetate carbon and oxygen atoms are respectively
colored orange and red (top of figure). (b) A topology diagram
of the novel FAH b-strand arrangement is shown. b Strands are
numbered in red according to their sequential occurrence in the
polypeptide chain; residue numbering is in black. Sheets A, B
and C are respectively colored in dark, light and medium shades
of blue, as in (a). a Helices are represented by red rectangles.
Figure 2, Figure 3 and Figure 4 and Figure 6b were generated
using MOLSCRIPT [40].
|
 |
|
|

|
| |
The above figure is
reprinted
by permission from Cell Press:
Structure
(1999,
7,
1023-1033)
copyright 1999.
|
|
| |
Figure was
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.Duhalde-Vega,
J.L.Aparicio,
and
L.A.Retegui
(2009).
Fine specificity of autoantibodies induced by mouse hepatitis virus A59.
|
| |
Viral Immunol,
22,
287-294.
|
 |
|
|
|
|
 |
S.Watanabe,
and
K.Makino
(2009).
Novel modified version of nonphosphorylated sugar metabolism--an alternative L-rhamnose pathway of Sphingomonas sp.
|
| |
FEBS J,
276,
1554-1567.
|
 |
|
|
|
|
 |
A.L.Fisher,
K.E.Page,
G.J.Lithgow,
and
L.Nash
(2008).
The Caenorhabditis elegans K10C2.4 gene encodes a member of the fumarylacetoacetate hydrolase family: a Caenorhabditis elegans model of type I tyrosinemia.
|
| |
J Biol Chem,
283,
9127-9135.
|
 |
|
|
|
|
 |
H.Mizutani,
and
N.Kunishima
(2007).
Purification, crystallization and preliminary X-ray analysis of the fumarylacetoacetase family member TTHA0809 from Thermus thermophilus HB8.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
63,
792-794.
|
 |
|
|
|
|
 |
M.De,
J.Bell,
N.J.Blackburn,
R.E.Mains,
and
B.A.Eipper
(2006).
Role for an essential tyrosine in peptide amidation.
|
| |
J Biol Chem,
281,
20873-20882.
|
 |
|
|
|
|
 |
N.Dreumont,
A.Maresca,
J.F.Boisclair-Lachance,
A.Bergeron,
and
R.M.Tanguay
(2005).
A minor alternative transcript of the fumarylacetoacetate hydrolase gene produces a protein despite being likely subjected to nonsense-mediated mRNA decay.
|
| |
BMC Mol Biol,
6,
1.
|
 |
|
|
|
|
 |
B.A.Manjasetty,
F.H.Niesen,
H.Delbrück,
F.Götz,
V.Sievert,
K.Büssow,
J.Behlke,
and
U.Heinemann
(2004).
X-ray structure of fumarylacetoacetate hydrolase family member Homo sapiens FLJ36880.
|
| |
Biol Chem,
385,
935-942.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
I.Fujii,
Y.Yasuoka,
H.F.Tsai,
Y.C.Chang,
K.J.Kwon-Chung,
and
Y.Ebizuka
(2004).
Hydrolytic polyketide shortening by ayg1p, a novel enzyme involved in fungal melanin biosynthesis.
|
| |
J Biol Chem,
279,
44613-44620.
|
 |
|
|
|
|
 |
N.Leulliot,
S.Quevillon-Cheruel,
I.Sorel,
M.Graille,
P.Meyer,
D.Liger,
K.Blondeau,
J.Janin,
and
H.van Tilbeurgh
(2004).
Crystal structure of yeast allantoicase reveals a repeated jelly roll motif.
|
| |
J Biol Chem,
279,
23447-23452.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
P.A.Mathieu,
K.A.Gómez,
J.P.Coutelier,
and
L.A.Retegui
(2004).
Sequence similarity and structural homologies are involved in the autoimmune response elicited by mouse hepatitis virus A59.
|
| |
J Autoimmun,
23,
117-126.
|
 |
|
|
|
|
 |
J.K.McIninch,
J.D.McIninch,
and
S.W.May
(2003).
Catalysis, stereochemistry, and inhibition of ureidoglycolate lyase.
|
| |
J Biol Chem,
278,
50091-50100.
|
 |
|
|
|
|
 |
J.A.Arranz,
F.Piñol,
L.Kozak,
C.Pérez-Cerdá,
B.Cormand,
M.Ugarte,
and
E.Riudor
(2002).
Splicing mutations, mainly IVS6-1(G>T), account for 70% of fumarylacetoacetate hydrolase (FAH) gene alterations, including 7 novel mutations, in a survey of 29 tyrosinemia type I patients.
|
| |
Hum Mutat,
20,
180-188.
|
 |
|
|
|
|
 |
J.R.Tame,
K.Namba,
E.J.Dodson,
and
D.I.Roper
(2002).
The crystal structure of HpcE, a bifunctional decarboxylase/isomerase with a multifunctional fold.
|
| |
Biochemistry,
41,
2982-2989.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.G.Levitt
(2001).
A new software routine that automates the fitting of protein X-ray crystallographic electron-density maps.
|
| |
Acta Crystallogr D Biol Crystallogr,
57,
1013-1019.
|
 |
|
|
|
|
 |
N.Dreumont,
J.A.Poudrier,
A.Bergeron,
H.L.Levy,
F.Baklouti,
and
R.M.Tanguay
(2001).
A missense mutation (Q279R) in the fumarylacetoacetate hydrolase gene, responsible for hereditary tyrosinemia, acts as a splicing mutation.
|
| |
BMC Genet,
2,
9.
|
 |
|
|
|
|
 |
S.E.Ealick
(2000).
Advances in multiple wavelength anomalous diffraction crystallography.
|
| |
Curr Opin Chem Biol,
4,
495-499.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

