 |
PDBsum entry 1psc

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.1.8.1
- aryldialkylphosphatase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
An aryl dialkyl phosphate + H2O = dialkyl phosphate + an aryl alcohol
|
 |
 |
 |
 |
 |
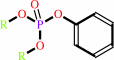 aryl dialkyl phosphate
aryl dialkyl phosphate
|
+
|
H2O
|
=
|
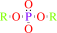 dialkyl phosphate
dialkyl phosphate
|
+
|
aryl alcohol
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Divalent cation
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
34:7973-7978
(1995)
|
|
PubMed id:
|
|
|
|

|
| |
|
Three-dimensional structure of the binuclear metal center of phosphotriesterase.
|
|
M.M.Benning,
J.M.Kuo,
F.M.Raushel,
H.M.Holden.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Phosphotriesterase, as isolated from Pseudomonas diminuta, is capable of
detoxifying widely used pesticides such as paraoxon and parathion and various
mammalian acetylcholinesterase inhibitors. The enzyme requires a binuclear metal
center for activity. Recently, the three-dimensional structure of the apoenzyme
was solved (Benning et al., 1994) and shown to consist of an alpha/beta-barrel.
Here we describe the three-dimensional structure of the holoenzyme,
reconstituted with cadmium, as determined by X-ray crystallographic analysis to
2.0-A resolution. Crystals employed in the investigation belonged to the space
group C2 with unit cell dimensions of a = 129.5 A, b = 91.4 A, c = 69.4 A, beta
= 91.9 degrees, and two subunits in the asymmetric unit. There are significant
differences in the three-dimensional architecture of the apo and holo forms of
the enzyme such that their alpha-carbon positions superimpose with a
root-mean-square deviation of 3.4 A. The binuclear metal center is located at
the C-terminus of the beta-barrel with the cadmiums separated by 3.8 A. There
are two bridging ligands to the metals: a water molecule (or possibly a
hydroxide ion) and a carbamylated lysine residue (Lys 169). The more buried
cadmium is surrounded by His 55, His 57, Lys 169, Asp 301, and the bridging
water in a trigonal bipyramidal arrangement. The second metal is coordinated in
a distorted octahedral geometry by His 201, His 230, Lys 169, the bridging water
molecule, and two additional solvents.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
C.M.Theriot,
and
A.M.Grunden
(2011).
Hydrolysis of organophosphorus compounds by microbial enzymes.
|
| |
Appl Microbiol Biotechnol,
89,
35-43.
|
 |
|
|
|
|
 |
L.Briseño-Roa,
C.M.Timperley,
A.D.Griffiths,
and
A.R.Fersht
(2011).
Phosphotriesterase variants with high methylphosphonatase activity and strong negative trade-off against phosphotriesters.
|
| |
Protein Eng Des Sel,
24,
151-159.
|
 |
|
|
|
|
 |
F.Ely,
K.S.Hadler,
L.R.Gahan,
L.W.Guddat,
D.L.Ollis,
and
G.Schenk
(2010).
The organophosphate-degrading enzyme from Agrobacterium radiobacter displays mechanistic flexibility for catalysis.
|
| |
Biochem J,
432,
565-573.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.L.Foo,
C.J.Jackson,
P.D.Carr,
H.K.Kim,
G.Schenk,
L.R.Gahan,
and
D.L.Ollis
(2010).
Mutation of outer-shell residues modulates metal ion co-ordination strength in a metalloenzyme.
|
| |
Biochem J,
429,
313-321.
|
 |
|
|
|
|
 |
B.Chen,
C.Lei,
Y.Shin,
and
J.Liu
(2009).
Probing mechanisms for enzymatic activity enhancement of organophosphorus hydrolase in functionalized mesoporous silica.
|
| |
Biochem Biophys Res Commun,
390,
1177-1181.
|
 |
|
|
|
|
 |
C.Y.Huang,
C.C.Hsu,
M.C.Chen,
and
Y.S.Yang
(2009).
Effect of metal binding and posttranslational lysine carboxylation on the activity of recombinant hydantoinase.
|
| |
J Biol Inorg Chem,
14,
111-121.
|
 |
|
|
|
|
 |
P.Gorla,
J.P.Pandey,
S.Parthasarathy,
M.Merrick,
and
D.Siddavattam
(2009).
Organophosphate hydrolase in Brevundimonas diminuta is targeted to the periplasmic face of the inner membrane by the twin arginine translocation pathway.
|
| |
J Bacteriol,
191,
6292-6299.
|
 |
|
|
|
|
 |
X.Zhang,
R.Wu,
L.Song,
Y.Lin,
M.Lin,
Z.Cao,
W.Wu,
and
Y.Mo
(2009).
Molecular dynamics simulations of the detoxification of paraoxon catalyzed by phosphotriesterase.
|
| |
J Comput Chem,
30,
2388-2401.
|
 |
|
|
|
|
 |
R.E.Mirams,
S.J.Smith,
K.S.Hadler,
D.L.Ollis,
G.Schenk,
and
L.R.Gahan
(2008).
Cadmium(II) complexes of the glycerophosphodiester-degrading enzyme GpdQ and a biomimetic N,O ligand.
|
| |
J Biol Inorg Chem,
13,
1065-1072.
|
 |
|
|
|
|
 |
Y.D.Champouret,
W.J.Nodes,
J.A.Scrimshire,
K.Singh,
G.A.Solan,
and
I.Young
(2007).
Sterically variable dizinc complexes bearing bis(iminopyridyl)phenolate ligands: synthesis, structures and reactivity studies.
|
| |
Dalton Trans,
(),
4565-4575.
|
 |
|
|
|
|
 |
M.Goto,
H.Hayashi,
I.Miyahara,
K.Hirotsu,
M.Yoshida,
and
T.Oikawa
(2006).
Crystal structures of nonoxidative zinc-dependent 2,6-dihydroxybenzoate (gamma-resorcylate) decarboxylase from Rhizobium sp. strain MTP-10005.
|
| |
J Biol Chem,
281,
34365-34373.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.K.Kim,
S.B.Mulrooney,
and
R.P.Hausinger
(2005).
Biosynthesis of active Bacillus subtilis urease in the absence of known urease accessory proteins.
|
| |
J Bacteriol,
187,
7150-7154.
|
 |
|
|
|
|
 |
J.Li,
J.B.Cross,
T.Vreven,
S.O.Meroueh,
S.Mobashery,
and
H.B.Schlegel
(2005).
Lysine carboxylation in proteins: OXA-10 beta-lactamase.
|
| |
Proteins,
61,
246-257.
|
 |
|
|
|
|
 |
L.Merone,
L.Mandrich,
M.Rossi,
and
G.Manco
(2005).
A thermostable phosphotriesterase from the archaeon Sulfolobus solfataricus: cloning, overexpression and properties.
|
| |
Extremophiles,
9,
297-305.
|
 |
|
|
|
|
 |
S.H.Nam,
H.S.Park,
and
H.S.Kim
(2005).
Evolutionary relationship and application of a superfamily of cyclic amidohydrolase enzymes.
|
| |
Chem Rec,
5,
298-307.
|
 |
|
|
|
|
 |
T.Liu,
A.A.Neverov,
J.S.Tsang,
and
R.S.Brown
(2005).
Mechanistic studies of La3+- and Zn2+-catalyzed methanolysis of aryl phosphate and phosphorothioate triesters. Development of artificial phosphotriesterase systems.
|
| |
Org Biomol Chem,
3,
1525-1533.
|
 |
|
|
|
|
 |
L.Sun,
Y.Dong,
Y.Zhou,
M.Yang,
C.Zhang,
Z.Rao,
and
X.E.Zhang
(2004).
Crystallization and preliminary X-ray studies of methyl parathion hydrolase from Pseudomonas sp. WBC-3.
|
| |
Acta Crystallogr D Biol Crystallogr,
60,
954-956.
|
 |
|
|
|
|
 |
P.R.Hall,
R.Zheng,
L.Antony,
M.Pusztai-Carey,
P.R.Carey,
and
V.C.Yee
(2004).
Transcarboxylase 5S structures: assembly and catalytic mechanism of a multienzyme complex subunit.
|
| |
EMBO J,
23,
3621-3631.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.C.Deo,
E.F.Schmidt,
A.Elhabazi,
H.Togashi,
S.K.Burley,
and
S.M.Strittmatter
(2004).
Structural bases for CRMP function in plexin-dependent semaphorin3A signaling.
|
| |
EMBO J,
23,
9.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Fokine,
R.Morales,
C.Contreras-Martel,
P.Carpentier,
F.Renault,
D.Rochu,
and
E.Chabriere
(2003).
Direct phasing at low resolution of a protein copurified with human paraoxonase (PON1).
|
| |
Acta Crystallogr D Biol Crystallogr,
59,
2083-2087.
|
 |
|
|
|
|
 |
R.Schwarzenbacher,
J.M.Canaves,
L.S.Brinen,
X.Dai,
A.M.Deacon,
M.A.Elsliger,
S.Eshaghi,
R.Floyd,
A.Godzik,
C.Grittini,
S.K.Grzechnik,
C.Guda,
L.Jaroszewski,
C.Karlak,
H.E.Klock,
E.Koesema,
J.S.Kovarik,
A.Kreusch,
P.Kuhn,
S.A.Lesley,
D.McMullan,
T.M.McPhillips,
M.A.Miller,
M.D.Miller,
A.Morse,
K.Moy,
J.Ouyang,
A.Robb,
K.Rodrigues,
T.L.Selby,
G.Spraggon,
R.C.Stevens,
H.van den Bedem,
J.Velasquez,
J.Vincent,
X.Wang,
B.West,
G.Wolf,
K.O.Hodgson,
J.Wooley,
and
I.A.Wilson
(2003).
Crystal structure of uronate isomerase (TM0064) from Thermotoga maritima at 2.85 A resolution.
|
| |
Proteins,
53,
142-145.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.B.Mulrooney,
and
R.P.Hausinger
(2003).
Metal ion dependence of recombinant Escherichia coli allantoinase.
|
| |
J Bacteriol,
185,
126-134.
|
 |
|
|
|
|
 |
Z.Gojkovic,
L.Rislund,
B.Andersen,
M.P.Sandrini,
P.F.Cook,
K.D.Schnackerz,
and
J.Piskur
(2003).
Dihydropyrimidine amidohydrolases and dihydroorotases share the same origin and several enzymatic properties.
|
| |
Nucleic Acids Res,
31,
1683-1692.
|
 |
|
|
|
|
 |
C.M.Cho,
A.Mulchandani,
and
W.Chen
(2002).
Bacterial cell surface display of organophosphorus hydrolase for selective screening of improved hydrolysis of organophosphate nerve agents.
|
| |
Appl Environ Microbiol,
68,
2026-2030.
|
 |
|
|
|
|
 |
D.Rochu,
and
P.Masson
(2002).
Multiple advantages of capillary zone electrophoresis for exploring protein conformational stability.
|
| |
Electrophoresis,
23,
189-202.
|
 |
|
|
|
|
 |
N.Shapir,
J.P.Osborne,
G.Johnson,
M.J.Sadowsky,
and
L.P.Wackett
(2002).
Purification, substrate range, and metal center of AtzC: the N-isopropylammelide aminohydrolase involved in bacterial atrazine metabolism.
|
| |
J Bacteriol,
184,
5376-5384.
|
 |
|
|
|
|
 |
J.A.Gerlt,
and
P.C.Babbitt
(2001).
Divergent evolution of enzymatic function: mechanistically diverse superfamilies and functionally distinct suprafamilies.
|
| |
Annu Rev Biochem,
70,
209-246.
|
 |
|
|
|
|
 |
J.L.Seffernick,
M.L.de Souza,
M.J.Sadowsky,
and
L.P.Wackett
(2001).
Melamine deaminase and atrazine chlorohydrolase: 98 percent identical but functionally different.
|
| |
J Bacteriol,
183,
2405-2410.
|
 |
|
|
|
|
 |
M.Chen-Goodspeed,
M.A.Sogorb,
F.Wu,
S.B.Hong,
and
F.M.Raushel
(2001).
Structural determinants of the substrate and stereochemical specificity of phosphotriesterase.
|
| |
Biochemistry,
40,
1325-1331.
|
 |
|
|
|
|
 |
M.M.Benning,
H.Shim,
F.M.Raushel,
and
H.M.Holden
(2001).
High resolution X-ray structures of different metal-substituted forms of phosphotriesterase from Pseudomonas diminuta.
|
| |
Biochemistry,
40,
2712-2722.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.Kimura
(2000).
Dimetallic hydrolases and their models.
|
| |
Curr Opin Chem Biol,
4,
207-213.
|
 |
|
|
|
|
 |
H.Shim,
and
F.M.Raushel
(2000).
Self-assembly of the binuclear metal center of phosphotriesterase.
|
| |
Biochemistry,
39,
7357-7364.
|
 |
|
|
|
|
 |
L.Maveyraud,
D.Golemi,
L.P.Kotra,
S.Tranier,
S.Vakulenko,
S.Mobashery,
and
J.P.Samama
(2000).
Insights into class D beta-lactamases are revealed by the crystal structure of the OXA10 enzyme from Pseudomonas aeruginosa.
|
| |
Structure,
8,
1289-1298.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
N.M.Okeley,
and
W.A.van der Donk
(2000).
Novel cofactors via post-translational modifications of enzyme active sites.
|
| |
Chem Biol,
7,
R159-R171.
|
 |
|
|
|
|
 |
S.Bartoschek,
J.A.Vorholt,
R.K.Thauer,
B.H.Geierstanger,
and
C.Griesinger
(2000).
N-carboxymethanofuran (carbamate) formation from methanofuran and CO2 in methanogenic archaea. Thermodynamics and kinetics of the spontaneous reaction.
|
| |
Eur J Biochem,
267,
3130-3138.
|
 |
|
|
|
|
 |
A.A.Morollo,
G.A.Petsko,
and
D.Ringe
(1999).
Structure of a Michaelis complex analogue: propionate binds in the substrate carboxylate site of alanine racemase.
|
| |
Biochemistry,
38,
3293-3301.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
B.diSioudi,
J.K.Grimsley,
K.Lai,
and
J.R.Wild
(1999).
Modification of near active site residues in organophosphorus hydrolase reduces metal stoichiometry and alters substrate specificity.
|
| |
Biochemistry,
38,
2866-2872.
|
 |
|
|
|
|
 |
J.G.Oakeshott,
C.Claudianos,
R.J.Russell,
and
G.C.Robin
(1999).
Carboxyl/cholinesterases: a case study of the evolution of a successful multigene family.
|
| |
Bioessays,
21,
1031-1042.
|
 |
|
|
|
|
 |
H.Shim,
S.B.Hong,
and
F.M.Raushel
(1998).
Hydrolysis of phosphodiesters through transformation of the bacterial phosphotriesterase.
|
| |
J Biol Chem,
273,
17445-17450.
|
 |
|
|
|
|
 |
J.A.Gerlt,
and
P.C.Babbitt
(1998).
Mechanistically diverse enzyme superfamilies: the importance of chemistry in the evolution of catalysis.
|
| |
Curr Opin Chem Biol,
2,
607-612.
|
 |
|
|
|
|
 |
M.A.Pearson,
R.A.Schaller,
L.O.Michel,
P.A.Karplus,
and
R.P.Hausinger
(1998).
Chemical rescue of Klebsiella aerogenes urease variants lacking the carbamylated-lysine nickel ligand.
|
| |
Biochemistry,
37,
6214-6220.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.M.Kuo,
M.Y.Chae,
and
F.M.Raushel
(1997).
Perturbations to the active site of phosphotriesterase.
|
| |
Biochemistry,
36,
1982-1988.
|
 |
|
|
|
|
 |
L.M.Watkins,
H.J.Mahoney,
J.K.McCulloch,
and
F.M.Raushel
(1997).
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase.
|
| |
J Biol Chem,
272,
25596-25601.
|
 |
|
|
|
|
 |
L.M.Watkins,
J.M.Kuo,
M.Chen-Goodspeed,
and
F.M.Raushel
(1997).
A combinatorial library for the binuclear metal center of bacterial phosphotriesterase.
|
| |
Proteins,
29,
553-561.
|
 |
|
|
|
|
 |
S.B.Hong,
L.S.Mullins,
H.Shim,
and
F.M.Raushel
(1997).
Mechanism-based inhibitors for the inactivation of the bacterial phosphotriesterase.
|
| |
Biochemistry,
36,
9022-9028.
|
 |
|
|
|
|
 |
A.Volbeda,
J.C.Fontecilla-Camps,
and
M.Frey
(1996).
Novel metal sites in protein structures.
|
| |
Curr Opin Struct Biol,
6,
804-812.
|
 |
|
|
|
|
 |
E.Jabri,
and
P.A.Karplus
(1996).
Structures of the Klebsiella aerogenes urease apoenzyme and two active-site mutants.
|
| |
Biochemistry,
35,
10616-10626.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
I.S.Park,
and
R.P.Hausinger
(1996).
Metal ion interaction with urease and UreD-urease apoproteins.
|
| |
Biochemistry,
35,
5345-5352.
|
 |
|
|
|
|
 |
J.L.Vanhooke,
M.M.Benning,
F.M.Raushel,
and
H.M.Holden
(1996).
Three-dimensional structure of the zinc-containing phosphotriesterase with the bound substrate analog diethyl 4-methylbenzylphosphonate.
|
| |
Biochemistry,
35,
6020-6025.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.M.Benning,
K.L.Taylor,
Liu R-Q,
G.Yang,
H.Xiang,
G.Wesenberg,
D.Dunaway-Mariano,
and
H.M.Holden
(1996).
Structure of 4-chlorobenzoyl coenzyme A dehalogenase determined to 1.8 A resolution: an enzyme catalyst generated via adaptive mutation.
|
| |
Biochemistry,
35,
8103-8109.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
N.O.Concha,
B.A.Rasmussen,
K.Bush,
and
O.Herzberg
(1996).
Crystal structure of the wide-spectrum binuclear zinc beta-lactamase from Bacteroides fragilis.
|
| |
Structure,
4,
823-836.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.B.Hong,
and
F.M.Raushel
(1996).
Metal-substrate interactions facilitate the catalytic activity of the bacterial phosphotriesterase.
|
| |
Biochemistry,
35,
10904-10912.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

