 |
PDBsum entry 1dpm

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.1.8.1
- aryldialkylphosphatase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
An aryl dialkyl phosphate + H2O = dialkyl phosphate + an aryl alcohol
|
 |
 |
 |
 |
 |
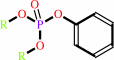 aryl dialkyl phosphate
aryl dialkyl phosphate
|
+
|
H2O
|
=
|
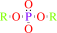 dialkyl phosphate
dialkyl phosphate
|
+
|
aryl alcohol
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Divalent cation
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
35:6020-6025
(1996)
|
|
PubMed id:
|
|
|
|

|
| |
|
Three-dimensional structure of the zinc-containing phosphotriesterase with the bound substrate analog diethyl 4-methylbenzylphosphonate.
|
|
J.L.Vanhooke,
M.M.Benning,
F.M.Raushel,
H.M.Holden.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Phosphotriesterase from Pseudomonas diminuta catalyzes the hydrolysis of
paraoxon and related acetylcholinesterase inhibitors with rate enhancements that
approach 10(12). The enzyme requires a binuclear metal center for activity and
as isolated contains 2 equiv of zinc per subunit. Here we describe the
three-dimensional structure of the Zn2+/Zn2+-substituted enzyme complexed with
the substrate analog diethyl 4-methylbenzylphosphonate. Crystals employed in the
investigation belonged to the space group C2 with unit cell dimensions of a =
129.6 A, b = 91.4 A, c = 69.4 A, beta = 91.9 degrees, and two subunits in the
asymmetric unit. The model was refined by least-squares analysis to a nominal
resolution of 2.1 A and a crystallographic R-factor of 15.4% for all measured
X-ray data. As in the previously reported structure of the cadmium-containing
enzyme, the bridging ligands are a carbamylated lysine residue (Lys 169) and a
hydroxide. The zinc ions are separated by 3.3 A. The more buried zinc ion is
surrounded by His 55, His 57, Lys 169, Asp 301, and the bridging hydroxide in a
trigonal bipyramidal arrangement as described for the cadmium-substituted
enzyme. Unlike the octahedral coordination observed for the more solvent-exposed
cadmium ion, however, the second zinc is tetrahedrally ligated to Lys 169, His
201, His 230, and the bridging hydroxide. The diethyl 4-methylbenzylphosphonate
occupies a site near the binuclear metal center with the phosphoryl oxygen of
the substrate analog situated at 3.5 A from the more solvent-exposed zinc ion.
The aromatic portion of the inhibitor binds in a fairly hydrophobic pocket. A
striking feature of the active site pocket is the lack of direct electrostatic
interactions between the inhibitor and the protein. This most likely explains
the broad substrate specificity exhibited by phosphotriesterase. The position of
the inhibitor within the active site suggests that the nucleophile for the
hydrolysis reaction is the metal-bound hydroxide.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
C.M.Theriot,
and
A.M.Grunden
(2011).
Hydrolysis of organophosphorus compounds by microbial enzymes.
|
| |
Appl Microbiol Biotechnol,
89,
35-43.
|
 |
|
|
|
|
 |
J.K.Raynes,
F.G.Pearce,
S.J.Meade,
and
J.A.Gerrard
(2011).
Immobilization of organophosphate hydrolase on an amyloid fibril nanoscaffold: Towards bioremediation and chemical detoxification.
|
| |
Biotechnol Prog,
27,
360-367.
|
 |
|
|
|
|
 |
L.Briseño-Roa,
C.M.Timperley,
A.D.Griffiths,
and
A.R.Fersht
(2011).
Phosphotriesterase variants with high methylphosphonatase activity and strong negative trade-off against phosphotriesters.
|
| |
Protein Eng Des Sel,
24,
151-159.
|
 |
|
|
|
|
 |
L.Briseño-Roa,
Z.Oliynyk,
C.M.Timperley,
A.D.Griffiths,
and
A.R.Fersht
(2011).
Highest paraoxonase turnover rate found in a bacterial phosphotriesterase variant.
|
| |
Protein Eng Des Sel,
24,
209-211.
|
 |
|
|
|
|
 |
F.Ely,
K.S.Hadler,
L.R.Gahan,
L.W.Guddat,
D.L.Ollis,
and
G.Schenk
(2010).
The organophosphate-degrading enzyme from Agrobacterium radiobacter displays mechanistic flexibility for catalysis.
|
| |
Biochem J,
432,
565-573.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
B.Chen,
C.Lei,
Y.Shin,
and
J.Liu
(2009).
Probing mechanisms for enzymatic activity enhancement of organophosphorus hydrolase in functionalized mesoporous silica.
|
| |
Biochem Biophys Res Commun,
390,
1177-1181.
|
 |
|
|
|
|
 |
X.Zhang,
R.Wu,
L.Song,
Y.Lin,
M.Lin,
Z.Cao,
W.Wu,
and
Y.Mo
(2009).
Molecular dynamics simulations of the detoxification of paraoxon catalyzed by phosphotriesterase.
|
| |
J Comput Chem,
30,
2388-2401.
|
 |
|
|
|
|
 |
A.B.Curtiss,
M.Bera,
G.T.Musie,
and
D.R.Powell
(2008).
Synthesis and characterization of mono- and micro6-sulfato hexanuclear zinc complexes of a new symmetric dinucleating ligand.
|
| |
Dalton Trans,
(),
2717-2724.
|
 |
|
|
|
|
 |
C.Neuhäuser,
D.Domide,
J.Mautz,
E.Kaifer,
and
H.J.Himmel
(2008).
Electron density controlled carbamate ligand binding mode: towards a better understanding of metalloenzyme activity.
|
| |
Dalton Trans,
(),
1821-1824.
|
 |
|
|
|
|
 |
R.E.Mirams,
S.J.Smith,
K.S.Hadler,
D.L.Ollis,
G.Schenk,
and
L.R.Gahan
(2008).
Cadmium(II) complexes of the glycerophosphodiester-degrading enzyme GpdQ and a biomimetic N,O ligand.
|
| |
J Biol Inorg Chem,
13,
1065-1072.
|
 |
|
|
|
|
 |
H.Jiang,
C.Yang,
H.Qu,
Z.Liu,
Q.S.Fu,
and
C.Qiao
(2007).
Cloning of a novel aldo-keto reductase gene from Klebsiella sp. strain F51-1-2 and its functional expression in Escherichia coli.
|
| |
Appl Environ Microbiol,
73,
4959-4965.
|
 |
|
|
|
|
 |
M.Elias,
J.Dupuy,
L.Merone,
C.Lecomte,
M.Rossi,
P.Masson,
G.Manco,
and
E.Chabriere
(2007).
Crystallization and preliminary X-ray diffraction analysis of the hyperthermophilic Sulfolobus solfataricus phosphotriesterase.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
63,
553-555.
|
 |
|
|
|
|
 |
E.Efremenko,
Y.Votchitseva,
F.Plieva,
I.Galaev,
and
B.Mattiasson
(2006).
Purification of His6-organophosphate hydrolase using monolithic supermacroporous polyacrylamide cryogels developed for immobilized metal affinity chromatography.
|
| |
Appl Microbiol Biotechnol,
70,
558-563.
|
 |
|
|
|
|
 |
I.Horne,
X.Qiu,
D.L.Ollis,
R.J.Russell,
and
J.G.Oakeshott
(2006).
Functional effects of amino acid substitutions within the large binding pocket of the phosphotriesterase OpdA from Agrobacterium sp. P230.
|
| |
FEMS Microbiol Lett,
259,
187-194.
|
 |
|
|
|
|
 |
C.J.Jackson,
J.W.Liu,
M.L.Coote,
and
D.L.Ollis
(2005).
The effects of substrate orientation on the mechanism of a phosphotriesterase.
|
| |
Org Biomol Chem,
3,
4343-4350.
|
 |
|
|
|
|
 |
F.J.Stevens,
C.Kuemmel,
G.Babnigg,
and
F.R.Collart
(2005).
Efficient recognition of protein fold at low sequence identity by conservative application of Psi-BLAST: application.
|
| |
J Mol Recognit,
18,
150-157.
|
 |
|
|
|
|
 |
L.L.Lin,
W.H.Hsu,
W.Y.Hsu,
S.C.Kan,
and
H.Y.Hu
(2005).
Phylogenetic analysis and biochemical characterization of a thermostable dihydropyrimidinase from alkaliphilic Bacillus sp. TS-23.
|
| |
Antonie Van Leeuwenhoek,
88,
189-197.
|
 |
|
|
|
|
 |
L.Sun,
Y.Dong,
Y.Zhou,
M.Yang,
C.Zhang,
Z.Rao,
and
X.E.Zhang
(2004).
Crystallization and preliminary X-ray studies of methyl parathion hydrolase from Pseudomonas sp. WBC-3.
|
| |
Acta Crystallogr D Biol Crystallogr,
60,
954-956.
|
 |
|
|
|
|
 |
A.D.Griffiths,
and
D.S.Tawfik
(2003).
Directed evolution of an extremely fast phosphotriesterase by in vitro compartmentalization.
|
| |
EMBO J,
22,
24-35.
|
 |
|
|
|
|
 |
I.Horne,
X.Qiu,
R.J.Russell,
and
J.G.Oakeshott
(2003).
The phosphotriesterase gene opdA in Agrobacterium radiobacter P230 is transposable.
|
| |
FEMS Microbiol Lett,
222,
1-8.
|
 |
|
|
|
|
 |
J.Koca,
C.G.Zhan,
R.C.Rittenhouse,
and
R.L.Ornstein
(2003).
Coordination number of zinc ions in the phosphotriesterase active site by molecular dynamics and quantum mechanics.
|
| |
J Comput Chem,
24,
368-378.
|
 |
|
|
|
|
 |
Z.Xu,
Y.Liu,
Y.Yang,
W.Jiang,
E.Arnold,
and
J.Ding
(2003).
Crystal structure of D-Hydantoinase from Burkholderia pickettii at a resolution of 2.7 Angstroms: insights into the molecular basis of enzyme thermostability.
|
| |
J Bacteriol,
185,
4038-4049.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.M.Cho,
A.Mulchandani,
and
W.Chen
(2002).
Bacterial cell surface display of organophosphorus hydrolase for selective screening of improved hydrolysis of organophosphate nerve agents.
|
| |
Appl Environ Microbiol,
68,
2026-2030.
|
 |
|
|
|
|
 |
D.Rochu,
and
P.Masson
(2002).
Multiple advantages of capillary zone electrophoresis for exploring protein conformational stability.
|
| |
Electrophoresis,
23,
189-202.
|
 |
|
|
|
|
 |
F.M.Raushel
(2002).
Bacterial detoxification of organophosphate nerve agents.
|
| |
Curr Opin Microbiol,
5,
288-295.
|
 |
|
|
|
|
 |
I.Horne,
T.D.Sutherland,
R.L.Harcourt,
R.J.Russell,
and
J.G.Oakeshott
(2002).
Identification of an opd (organophosphate degradation) gene in an Agrobacterium isolate.
|
| |
Appl Environ Microbiol,
68,
3371-3376.
|
 |
|
|
|
|
 |
D.Walther,
C.Fugger,
H.Schreer,
R.Kilian,
and
H.Görls
(2001).
Reversible fixation of carbon dioxide at nickel(0) centers: a route for large organometallic rings, dimers, and tetramers.
|
| |
Chemistry,
7,
5214-5221.
|
 |
|
|
|
|
 |
E.I.Scharff,
J.Koepke,
G.Fritzsch,
C.Lücke,
and
H.Rüterjans
(2001).
Crystal structure of diisopropylfluorophosphatase from Loligo vulgaris.
|
| |
Structure,
9,
493-502.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.A.Gerlt,
and
P.C.Babbitt
(2001).
Divergent evolution of enzymatic function: mechanistically diverse superfamilies and functionally distinct suprafamilies.
|
| |
Annu Rev Biochem,
70,
209-246.
|
 |
|
|
|
|
 |
J.Hartleib,
and
H.Rüterjans
(2001).
Insights into the reaction mechanism of the diisopropyl fluorophosphatase from Loligo vulgaris by means of kinetic studies, chemical modification and site-directed mutagenesis.
|
| |
Biochim Biophys Acta,
1546,
312-324.
|
 |
|
|
|
|
 |
M.Chen-Goodspeed,
M.A.Sogorb,
F.Wu,
and
F.M.Raushel
(2001).
Enhancement, relaxation, and reversal of the stereoselectivity for phosphotriesterase by rational evolution of active site residues.
|
| |
Biochemistry,
40,
1332-1339.
|
 |
|
|
|
|
 |
M.Chen-Goodspeed,
M.A.Sogorb,
F.Wu,
S.B.Hong,
and
F.M.Raushel
(2001).
Structural determinants of the substrate and stereochemical specificity of phosphotriesterase.
|
| |
Biochemistry,
40,
1325-1331.
|
 |
|
|
|
|
 |
M.M.Benning,
H.Shim,
F.M.Raushel,
and
H.M.Holden
(2001).
High resolution X-ray structures of different metal-substituted forms of phosphotriesterase from Pseudomonas diminuta.
|
| |
Biochemistry,
40,
2712-2722.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.P.Pang
(2001).
Successful molecular dynamics simulation of two zinc complexes bridged by a hydroxide in phosphotriesterase using the cationic dummy atom method.
|
| |
Proteins,
45,
183-189.
|
 |
|
|
|
|
 |
H.Shim,
and
F.M.Raushel
(2000).
Self-assembly of the binuclear metal center of phosphotriesterase.
|
| |
Biochemistry,
39,
7357-7364.
|
 |
|
|
|
|
 |
B.diSioudi,
J.K.Grimsley,
K.Lai,
and
J.R.Wild
(1999).
Modification of near active site residues in organophosphorus hydrolase reduces metal stoichiometry and alters substrate specificity.
|
| |
Biochemistry,
38,
2866-2872.
|
 |
|
|
|
|
 |
E.Meyer,
T.J.Kappock,
C.Osuji,
and
J.Stubbe
(1999).
Evidence for the direct transfer of the carboxylate of N5-carboxyaminoimidazole ribonucleotide (N5-CAIR) to generate 4-carboxy-5-aminoimidazole ribonucleotide catalyzed by Escherichia coli PurE, an N5-CAIR mutase.
|
| |
Biochemistry,
38,
3012-3018.
|
 |
|
|
|
|
 |
S.B.Hong,
and
F.M.Raushel
(1999).
Stereochemical constraints on the substrate specificity of phosphotriesterase.
|
| |
Biochemistry,
38,
1159-1165.
|
 |
|
|
|
|
 |
S.Benini,
W.R.Rypniewski,
K.S.Wilson,
S.Miletti,
S.Ciurli,
and
S.Mangani
(1999).
A new proposal for urease mechanism based on the crystal structures of the native and inhibited enzyme from Bacillus pasteurii: why urea hydrolysis costs two nickels.
|
| |
Structure,
7,
205-216.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.Shim,
S.B.Hong,
and
F.M.Raushel
(1998).
Hydrolysis of phosphodiesters through transformation of the bacterial phosphotriesterase.
|
| |
J Biol Chem,
273,
17445-17450.
|
 |
|
|
|
|
 |
R.J.Kazlauskas,
and
H.K.Weber
(1998).
Improving hydrolases for organic synthesis.
|
| |
Curr Opin Chem Biol,
2,
121-126.
|
 |
|
|
|
|
 |
S.M.Fabiane,
M.K.Sohi,
T.Wan,
D.J.Payne,
J.H.Bateson,
T.Mitchell,
and
B.J.Sutton
(1998).
Crystal structure of the zinc-dependent beta-lactamase from Bacillus cereus at 1.9 A resolution: binuclear active site with features of a mononuclear enzyme.
|
| |
Biochemistry,
37,
12404-12411.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
U.Ermler,
W.Grabarse,
S.Shima,
M.Goubeaud,
and
R.K.Thauer
(1998).
Active sites of transition-metal enzymes with a focus on nickel.
|
| |
Curr Opin Struct Biol,
8,
749-758.
|
 |
|
|
|
|
 |
J.M.Kuo,
M.Y.Chae,
and
F.M.Raushel
(1997).
Perturbations to the active site of phosphotriesterase.
|
| |
Biochemistry,
36,
1982-1988.
|
 |
|
|
|
|
 |
L.M.Watkins,
H.J.Mahoney,
J.K.McCulloch,
and
F.M.Raushel
(1997).
Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase.
|
| |
J Biol Chem,
272,
25596-25601.
|
 |
|
|
|
|
 |
S.B.Hong,
L.S.Mullins,
H.Shim,
and
F.M.Raushel
(1997).
Mechanism-based inhibitors for the inactivation of the bacterial phosphotriesterase.
|
| |
Biochemistry,
36,
9022-9028.
|
 |
|
|
|
|
 |
S.Ohuchi,
H.Nakamura,
H.Sligiura,
M.Narita,
and
K.Sode
(1997).
An optical resolution of racemic organophosphorous esters by phosphotriesterase-catalyzing hydrolysis.
|
| |
Appl Biochem Biotechnol,
63,
659-665.
|
 |
|
|
|
|
 |
J.B.Thoden,
P.A.Frey,
and
H.M.Holden
(1996).
High-resolution X-ray structure of UDP-galactose 4-epimerase complexed with UDP-phenol.
|
| |
Protein Sci,
5,
2149-2161.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.B.Hong,
and
F.M.Raushel
(1996).
Metal-substrate interactions facilitate the catalytic activity of the bacterial phosphotriesterase.
|
| |
Biochemistry,
35,
10904-10912.
|
 |
|
|
|
|
 |
S.J.Cooper,
G.A.Leonard,
S.M.McSweeney,
A.W.Thompson,
J.H.Naismith,
S.Qamar,
A.Plater,
A.Berry,
and
W.N.Hunter
(1996).
The crystal structure of a class II fructose-1,6-bisphosphate aldolase shows a novel binuclear metal-binding active site embedded in a familiar fold.
|
| |
Structure,
4,
1303-1315.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

