Assemblies
Assembly Name:
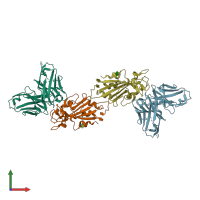
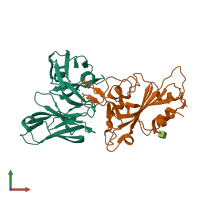
Spike protein S2' and IG-heavy chain and IG-heavy chain
Multimeric state:
hetero dimer
Accessible surface area:
18412.97 Å2
Buried surface area:
2014.26 Å2
Dissociation area:
867.05
Å2
Dissociation energy (ΔGdiss):
-1.16
kcal/mol
Dissociation entropy (TΔSdiss):
12.33
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-235060
Assembly Name:
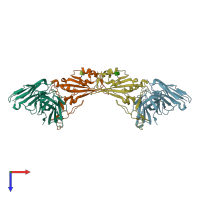
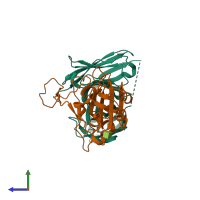
Spike protein S2' and IG-heavy chain and IG-heavy chain
Multimeric state:
hetero dimer
Accessible surface area:
18463.14 Å2
Buried surface area:
2074.61 Å2
Dissociation area:
895.23
Å2
Dissociation energy (ΔGdiss):
-1.4
kcal/mol
Dissociation entropy (TΔSdiss):
12.33
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-235060
Macromolecules
Chains: A, C
Length: 251 amino acids
Theoretical weight: 26.38 KDa
Source organism: Homo sapiens
Expression system: Drosophila melanogaster
Length: 251 amino acids
Theoretical weight: 26.38 KDa
Source organism: Homo sapiens
Expression system: Drosophila melanogaster
Chains: B, D
Length: 199 amino acids
Theoretical weight: 22.36 KDa
Source organism: Severe acute respiratory syndrome coronavirus 2
Expression system: Drosophila melanogaster
UniProt:
Pfam: Betacoronavirus spike glycoprotein S1, receptor binding
Length: 199 amino acids
Theoretical weight: 22.36 KDa
Source organism: Severe acute respiratory syndrome coronavirus 2
Expression system: Drosophila melanogaster
UniProt:
- Canonical:
 P0DTC2 (Residues: 334-527; Coverage: 15%)
P0DTC2 (Residues: 334-527; Coverage: 15%)
Pfam: Betacoronavirus spike glycoprotein S1, receptor binding