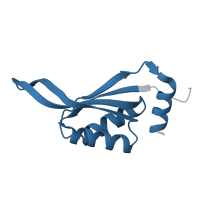
Crystal structure of E.coli CutA1 E61V/C16A/C39A/C79A mutation
Source organism:
Escherichia coli K-12
Biochemical function:
metal ion binding
Biological process: response to copper ion
Cellular component: protein-containing complex
Biological process: response to copper ion
Cellular component: protein-containing complex