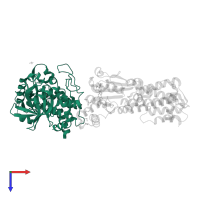
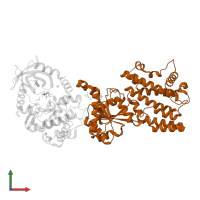
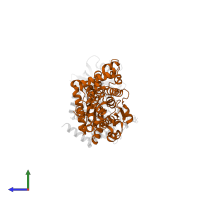
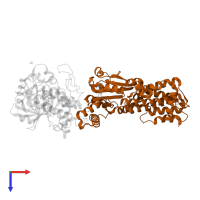
Assemblies
Multimeric state:
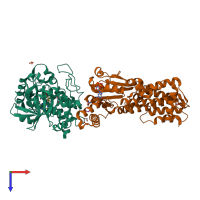
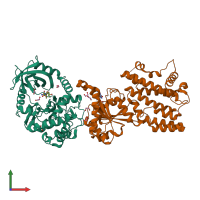
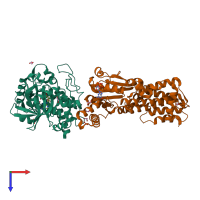
hetero dimer
Accessible surface area:
32884.41 Å2
Buried surface area:
5861.72 Å2
Dissociation area:
1,308.12
Å2
Dissociation energy (ΔGdiss):
-13.48
kcal/mol
Dissociation entropy (TΔSdiss):
13.67
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-125220
Assembly Name:
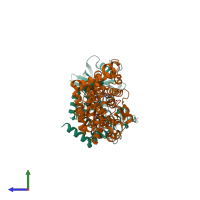
Protein kinase domain-containing protein
Multimeric state:
monomeric
Accessible surface area:
15974.77 Å2
Buried surface area:
1973.89 Å2
Dissociation area:
101.03
Å2
Dissociation energy (ΔGdiss):
-2.54
kcal/mol
Dissociation entropy (TΔSdiss):
-0.41
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-125219
Assembly Name:
Interferon-inducible GTPase 1
Multimeric state:
monomeric
Accessible surface area:
19030.48 Å2
Buried surface area:
1766.99 Å2
Dissociation area:
93.52
Å2
Dissociation energy (ΔGdiss):
-2.9
kcal/mol
Dissociation entropy (TΔSdiss):
-0.27
kcal/mol
Symmetry number:
1
PDBe Complex ID:
PDB-CPX-193063
Macromolecules
Chain: A
Length: 371 amino acids
Theoretical weight: 41.35 KDa
Source organism: Toxoplasma gondii type I
Expression system: Escherichia coli BL21(DE3)
UniProt:
Pfam: Kinase-like
InterPro:
Length: 371 amino acids
Theoretical weight: 41.35 KDa
Source organism: Toxoplasma gondii type I
Expression system: Escherichia coli BL21(DE3)
UniProt:
- Canonical:
 I6ZQR7 (Residues: 175-541; Coverage: 70%)
I6ZQR7 (Residues: 175-541; Coverage: 70%)
Pfam: Kinase-like
InterPro:
- Protein kinase-like domain, Apicomplexa
- Protein kinase-like domain superfamily
- Protein kinase domain
Chain: B
Length: 423 amino acids
Theoretical weight: 48.48 KDa
Source organism: Mus musculus
Expression system: Escherichia coli BL21(DE3)
UniProt:
Pfam: Interferon-inducible GTPase (IIGP)
InterPro:
Length: 423 amino acids
Theoretical weight: 48.48 KDa
Source organism: Mus musculus
Expression system: Escherichia coli BL21(DE3)
UniProt:
- Canonical:
 Q9QZ85 (Residues: 1-413; Coverage: 100%)
Q9QZ85 (Residues: 1-413; Coverage: 100%)
Pfam: Interferon-inducible GTPase (IIGP)
InterPro:
- P-loop containing nucleoside triphosphate hydrolase
- Immunity-related GTPases-like
- IRG-type guanine nucleotide-binding (G) domain