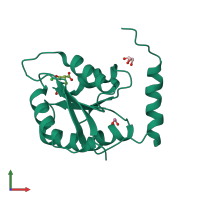
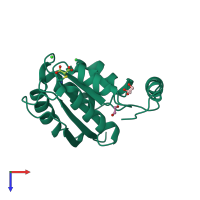
Assemblies
Assembly Name:
Putative ribose 5-phosphate isomerase
Multimeric state:
homo dimer
Accessible surface area:
11896.06 Å2
Buried surface area:
5826.32 Å2
Dissociation area:
2,062.06
Å2
Dissociation energy (ΔGdiss):
31.97
kcal/mol
Dissociation entropy (TΔSdiss):
12.42
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-143525
Assembly Name:
Putative ribose 5-phosphate isomerase
Multimeric state:
homo tetramer
Accessible surface area:
21149.67 Å2
Buried surface area:
14045.71 Å2
Dissociation area:
185.71
Å2
Dissociation energy (ΔGdiss):
28.31
kcal/mol
Dissociation entropy (TΔSdiss):
-1.64
kcal/mol
Symmetry number:
4
PDBe Complex ID:
PDB-CPX-143526
Macromolecules
Chain: A
Length: 184 amino acids
Theoretical weight: 19.71 KDa
Source organism: Coccidioides immitis RS
Expression system: Escherichia coli
UniProt:
Pfam: Ribose/Galactose Isomerase
InterPro:
Length: 184 amino acids
Theoretical weight: 19.71 KDa
Source organism: Coccidioides immitis RS
Expression system: Escherichia coli
UniProt:
- Canonical:
 P0CL19 (Residues: 1-163; Coverage: 100%)
P0CL19 (Residues: 1-163; Coverage: 100%)
Pfam: Ribose/Galactose Isomerase
InterPro:
- Sugar-phosphate isomerase, RpiB/LacA/LacB family
- Ribose 5-phosphate isomerase B, Actinobacteria-type
- Sugar-phosphate isomerase, RpiB/LacA/LacB superfamily