Introduction¶
Hint
Click here to go back to the SAT.
The Segmentation Annotation Tool (SAT) provides a simple way to richly annotate the EMDB-SFF files against ontologies and archives. Annotation is a way for users to add value to their hard-won segmentations by using stable, curated and reliable accessions to describe the components of a biological entity.
EMDB-SFF is an open segmentation data model created following community consultation. It is designed primarily for images derived from various electron microscopy modalities and captures segmentations using several geometrical primitives. In addition to representing segmentation geometry, EMDB-SFF is designed to capture annotations both in free text and fixed text through accessions from various resources such as ontologies and archives. The SAT provides a simple graphical user interface alternative to carry out annotation.
This guide outlines the steps you can take prior to and after annotating your segmentation to produce annotated EMDB-SFF files of your segmentations, which can then be deposited alongside your image data into EMDB and EMPIAR. It illustrates how to prepare various application-specific segmentation file formats so as to save both your time and disk space.
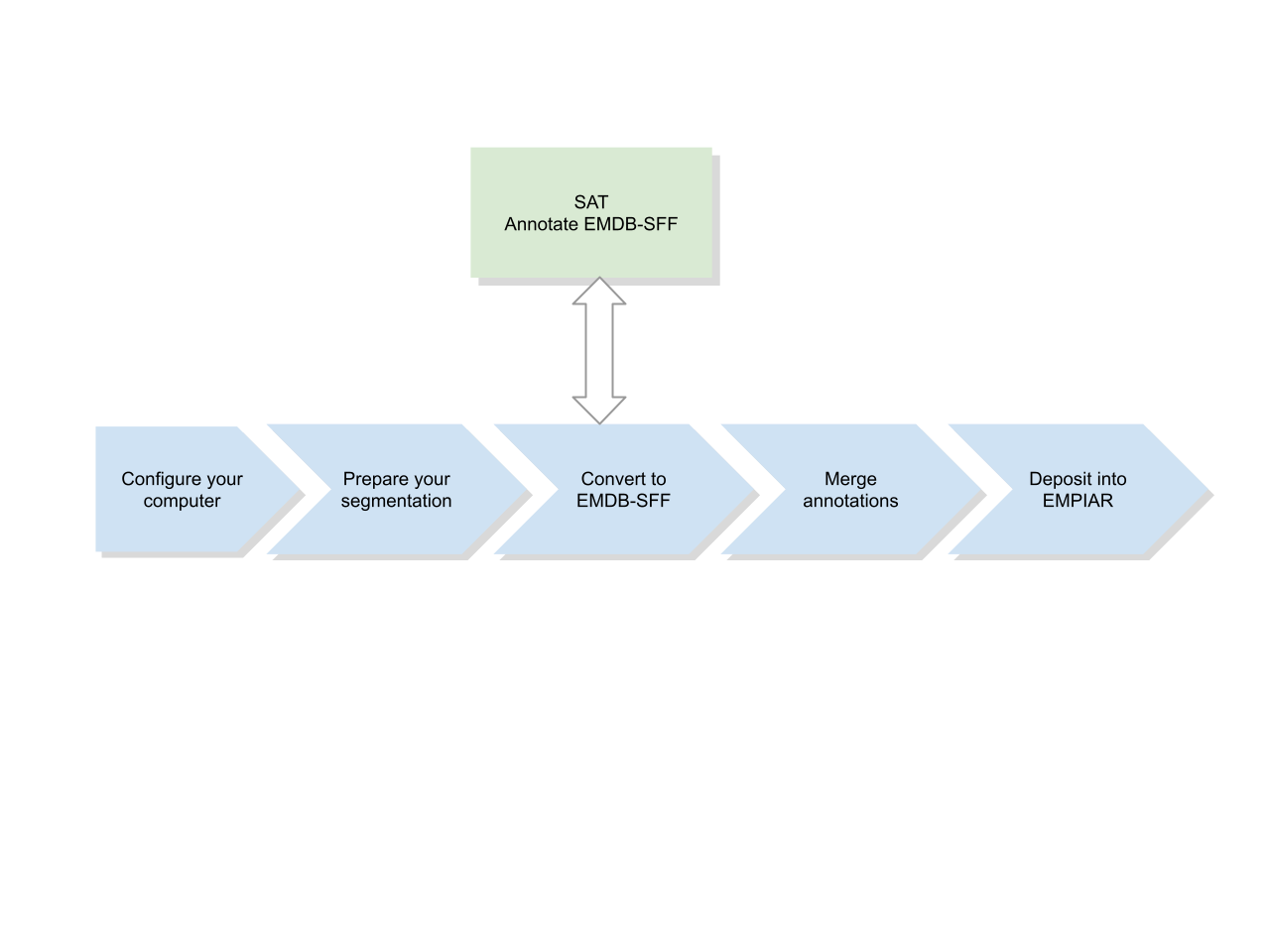
Install sfftk locally, prepare your segmentation then convert it to EMDB-SFF. The geometry can then be extracted to make a smaller JSON file to work with SAT. After annotation with SAT, download, merge then deposited your annotated EMDB-SFF file.¶