 |
PDBsum entry 4ogg
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase
|
 |
|
Title:
|
 |
Crystal structure of arabidopsis thaliana dj-1d with glyoxylate as substrate analog
|
|
Structure:
|
 |
Protein dj-1 homolog d. Chain: a, b, c. Synonym: atdj1d, lactoylglutathione lyase dj1d. Engineered: yes
|
|
Source:
|
 |
Arabidopsis thaliana. Mouse-ear cress. Organism_taxid: 3702. Gene: dj1d, at3g02720, f13e7.34. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
1.60Å
|
R-factor:
|
0.185
|
R-free:
|
0.212
|
|
|
Authors:
|
 |
D.Choi,J.Kim,K.-S.Ryu,C.Park
|
|
Key ref:
|
 |
D.Choi
et al.
(2014).
Stereospecific mechanism of DJ-1 glyoxalases inferred from their hemithioacetal-containing crystal structures.
Febs J,
281,
5447-5462.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
16-Jan-14
|
Release date:
|
15-Oct-14
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q9M8R4
(DJ1D_ARATH) -
Protein DJ-1 homolog D from Arabidopsis thaliana
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
388 a.a.
386 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 2 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.4.1.5
- lactoylglutathione lyase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(R)-S-lactoylglutathione = methylglyoxal + glutathione
|
 |
 |
 |
 |
 |
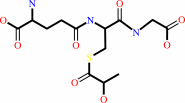 (R)-S-lactoylglutathione
(R)-S-lactoylglutathione
|
=
|
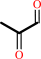 methylglyoxal
methylglyoxal
|
+
|
 glutathione
glutathione
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Febs J
281:5447-5462
(2014)
|
|
PubMed id:
|
|
|
|

|
| |
|
Stereospecific mechanism of DJ-1 glyoxalases inferred from their hemithioacetal-containing crystal structures.
|
|
D.Choi,
J.Kim,
S.Ha,
K.Kwon,
E.H.Kim,
H.Y.Lee,
K.S.Ryu,
C.Park.
|
|
|

|
| |
ABSTRACT
|
|

|
| |
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

