 |
PDBsum entry 2ayl

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
2ayl
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
2.0 angstrom crystal structure of manganese protoporphyrin ix- reconstituted ovine prostaglandin h2 synthase-1 complexed with flurbiprofen
|
|
Structure:
|
 |
Prostaglandin g/h synthase 1. Chain: a, b. Synonym: cyclooxygenase-1, cox-1, prostaglandin-endoperoxide synthase 1, prostaglandin h2 synthase 1, pgh synthase 1, pghs-1, phs 1. Ec: 1.14.99.1
|
|
Source:
|
 |
Ovis aries. Sheep. Organism_taxid: 9940. Organ: seminal vesicle
|
|
Biol. unit:
|
 |
Dimer (from
)
|
|
Resolution:
|
 |
|
2.00Å
|
R-factor:
|
0.218
|
R-free:
|
0.237
|
|
|
Authors:
|
 |
K.Gupta,B.S.Selinsky,P.J.Loll
|
Key ref:

|
 |
K.Gupta
et al.
(2006).
2.0 angstroms structure of prostaglandin H2 synthase-1 reconstituted with a manganese porphyrin cofactor.
Acta Crystallogr D Biol Crystallogr,
62,
151-156.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
07-Sep-05
|
Release date:
|
24-Jan-06
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P05979
(PGH1_SHEEP) -
Prostaglandin G/H synthase 1 from Ovis aries
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
600 a.a.
553 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 2 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.14.99.1
- prostaglandin-endoperoxide synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(5Z,8Z,11Z,14Z)-eicosatetraenoate + AH2 + 2 O2 = prostaglandin H2 + A + H2O
|
 |
 |
 |
 |
 |
(5Z,8Z,11Z,14Z)-eicosatetraenoate
|
+
|
AH2
|
+
|
 2
×
O2
2
×
O2
|
=
|
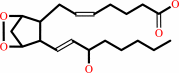 prostaglandin H2
prostaglandin H2
|
+
|
|
+
|
H2O
Bound ligand (Het Group name = )
matches with 51.11% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Acta Crystallogr D Biol Crystallogr
62:151-156
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
2.0 angstroms structure of prostaglandin H2 synthase-1 reconstituted with a manganese porphyrin cofactor.
|
|
K.Gupta,
B.S.Selinsky,
P.J.Loll.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Prostaglandin H2 synthase (EC 1.14.99.1) is a clinically important drug target
that catalyzes two key steps in the biosynthesis of the eicosanoid hormones. The
enzyme contains spatially distinct cyclooxygenase and peroxidase active sites,
both of which require a heme cofactor. Substitution of ferric heme by Mn(III)
protoporphyrin IX greatly diminishes the peroxidase activity, but has little
effect on the cyclooxygenase activity. Here, the 2.0 angstroms resolution
crystal structure of the Mn(III) form of ovine prostaglandin H2 synthase-1 is
described (R = 21.8%, R(free) = 23.7%). Substitution of Mn(III) for Fe(III)
causes no structural perturbations in the protein. However, the out-of-plane
displacement of the manganese ion with respect to the porphyrin is greater than
that of the iron by approximately 0.2 angstroms. This perturbation may help to
explain the altered catalytic properties of the manganese enzyme.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |
Figure 3.
Figure 3
Stereoviews of the peroxidase and cyclooxygenase active sites. (a) Superposition of the
peroxidase sites of the Fe-PGHS and Mn-PGHS structures. The iron structure is shown in red
and the manganese structure in blue. The protein backbone is represented by the yellow
ribbon. (b) The Mn-PGHS structure, showing the anomalous difference Fourier map contoured
at 6 [89][sigma] . The perspective of the viewer is similar to that in (a), but in (b) the
porphyrin has been rotated slightly upward about a horizontal axis relative to the
orientation shown in (a). (c) Superposition of the NSAID-binding sites of the 2.0 Å
Fe-PGHS and Mn-PGHS structures (Gupta et al., 2004[90] [Gupta, K., Selinsky, B. S., Kaub,
C. J., Katz, A. K. & Loll, P. J. (2004). J. Mol. Biol. 335, 503-518.]-[91][bluearr.gif]
and this work); the color scheme is the same as in (a). The ligand in the Fe-PGHS
structure is [92][alpha] -methyl-4-biphenylacetic acid (desfluoro-flurbiprofen). The
flurbiprofen in the Mn-PGHS structure occupies two alternate positions of roughly equal
occupancy, corresponding to two alternate positions for the F atom. The two fluorine
positions are marked by asterisks. Two alternate rotamers are observed for the side chain
of Ser530 in both the Fe- and Mn-PGHS structures.
|
 |
|
|

|
| |
The above figure is
reprinted
by permission from the IUCr:
Acta Crystallogr D Biol Crystallogr
(2006,
62,
151-156)
copyright 2006.
|
|
| |
Figure was
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.M.Lü,
C.E.Rogge,
G.Wu,
R.J.Kulmacz,
W.A.van der Donk,
and
A.L.Tsai
(2011).
Cyclooxygenase reaction mechanism of PGHS--evidence for a reversible transition between a pentadienyl radical and a new tyrosyl radical by nitric oxide trapping.
|
| |
J Inorg Biochem,
105,
356-365.
|
 |
|
|
|
|
 |
A.L.Tsai,
and
R.J.Kulmacz
(2010).
Prostaglandin H synthase: resolved and unresolved mechanistic issues.
|
| |
Arch Biochem Biophys,
493,
103-124.
|
 |
|
|
|
|
 |
J.Rand Doyen,
N.Yucer,
L.M.Lichtenberger,
and
R.J.Kulmacz
(2008).
Phospholipid actions on PGHS-1 and -2 cyclooxygenase kinetics.
|
| |
Prostaglandins Other Lipid Mediat,
85,
134-143.
|
 |
|
|
|
|
 |
T.Majtan,
L.R.Singh,
L.Wang,
W.D.Kruger,
and
J.P.Kraus
(2008).
Active Cystathionine {beta}-Synthase Can Be Expressed in Heme-free Systems in the Presence of Metal-substituted Porphyrins or a Chemical Chaperone.
|
| |
J Biol Chem,
283,
34588-34595.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

