 |
PDBsum entry 1v2e

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.6.1.15
- glutamine--pyruvate transaminase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-glutamine + pyruvate = 2-oxoglutaramate + L-alanine
|
 |
 |
 |
 |
 |
 L-glutamine
L-glutamine
|
+
|
pyruvate
Bound ligand (Het Group name = )
matches with 66.67% similarity
|
=
|
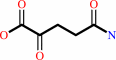 2-oxoglutaramate
2-oxoglutaramate
|
+
|
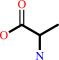 L-alanine
L-alanine
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
PLP)
matches with 93.75% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
279:16518-16525
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structures of glutamine:phenylpyruvate aminotransferase from Thermus thermophilus HB8: induced fit and substrate recognition.
|
|
M.Goto,
R.Omi,
I.Miyahara,
A.Hosono,
H.Mizuguchi,
H.Hayashi,
H.Kagamiyama,
K.Hirotsu.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The following three-dimensional structures of three forms of
glutamine:phenylpyruvate aminotransferase from Thermus thermophilus HB8 have
been determined and represent the first x-ray analysis of the enzyme: the
unliganded pyridoxal 5'-phosphate form at 1.9 A resolution and two complexes
with 3-phenylpropionate and alpha-keto-gamma-methylthiobutyrate at 2.35 and 2.6
A resolution, respectively. The enzyme shows high activity toward phenylalanine,
tyrosine, tryptophan, kynurenine, methionine, and glutamine. The enzyme is a
homodimer, and each subunit is divided into an N-terminal arm and small and
large domains. Based on its folding, the enzyme belongs to fold type I,
aminotransferase subclass Ib. The subclass I aminotransferases whose structures
have so far been determined exhibit a large movement of the small domain region
upon binding of a substrate. Similarly, the glutamine:phenylpyruvate
aminotransferase undergoes a large movement in part of the small domain to close
the active site. The active-site pocket has a shape and size suitable to enclose
the side chain of an aromatic amino acid or that of methionine. The inner side
of the pocket is mostly hydrophobic, but also has polar sites. The kynurenine
complex generated by computer modeling fits the pocket of the enzyme and its
hydrophilic groups interact with the polar sites of the pocket.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 5.
FIG. 5. Schematic diagram showing hydrogen bond and salt
bridge interactions of the active-site residues. Putative
interactions are shown by dotted lines if the acceptor and donor
are less than 3.5 Å apart. Phe-112 and Val-193, which
sandwich the pyridine ring of PLP, are omitted for clarity. A,
the unliganded ttGlnAT in the closed form. B, the ttGlnAT
complex with 3PP. The hydrogen bonds between Ser-13 and Asp-113
have been omitted for clarity.
|
 |
Figure 6.
FIG. 6. Superimposition of the active site of the
ttGlnAT·kynurenine complex model onto ttGlnAT·3PP.
The residues that represent the ttGlnAT· kynurenine
complex and ttGlnAT·3PP are shown in brown and deep blue,
respectively. Kynurenine and 3PP are represented by thick lines
in pale blue and brown. The oxygen and nitrogen atoms of
kynurenine and 3PP are colored red and deep blue, respectively.
The dotted lines (green) show the hydrogen bonding interactions
of the amino and carbonyl groups of the 3-anthraniloyl moiety of
kynurenine with Asp-113 and Tyr-57^*, respectively.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2004,
279,
16518-16525)
copyright 2004.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
Q.Han,
T.Cai,
D.A.Tagle,
and
J.Li
(2010).
Thermal stability, pH dependence and inhibition of four murine kynurenine aminotransferases.
|
| |
BMC Biochem,
11,
19.
|
 |
|
|
|
|
 |
Q.Han,
T.Cai,
D.A.Tagle,
and
J.Li
(2010).
Structure, expression, and function of kynurenine aminotransferases in human and rodent brains.
|
| |
Cell Mol Life Sci,
67,
353-368.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Q.Han,
H.Robinson,
T.Cai,
D.A.Tagle,
and
J.Li
(2009).
Biochemical and structural properties of mouse kynurenine aminotransferase III.
|
| |
Mol Cell Biol,
29,
784-793.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Q.Han,
H.Robinson,
T.Cai,
D.A.Tagle,
and
J.Li
(2009).
Structural insight into the inhibition of human kynurenine aminotransferase I/glutamine transaminase K.
|
| |
J Med Chem,
52,
2786-2793.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Q.Han,
Y.G.Gao,
H.Robinson,
and
J.Li
(2008).
Structural insight into the mechanism of substrate specificity of aedes kynurenine aminotransferase.
|
| |
Biochemistry,
47,
1622-1630.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.Chon,
H.Matsumura,
S.Shimizu,
N.Maeda,
Y.Koga,
K.Takano,
and
S.Kanaya
(2005).
Overproduction and preliminary crystallographic study of a human kynurenine aminotransferase II homologue from Pyrococcus horikoshii OT3.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
61,
319-322.
|
 |
|
|
|
|
 |
H.Chon,
H.Matsumura,
Y.Koga,
K.Takano,
and
S.Kanaya
(2005).
Crystal structure of a human kynurenine aminotransferase II homologue from Pyrococcus horikoshii OT3 at 2.20 A resolution.
|
| |
Proteins,
61,
685-688.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.Hirotsu,
M.Goto,
A.Okamoto,
and
I.Miyahara
(2005).
Dual substrate recognition of aminotransferases.
|
| |
Chem Rec,
5,
160-172.
|
 |
|
|
|
|
 |
Q.Han,
Y.G.Gao,
H.Robinson,
H.Ding,
S.Wilson,
and
J.Li
(2005).
Crystal structures of Aedes aegypti kynurenine aminotransferase.
|
| |
FEBS J,
272,
2198-2206.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

