 |
PDBsum entry 3e2f

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Transferase, lyase
|
PDB id
|
|

|

|
3e2f
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase, lyase
|
 |
|
Title:
|
 |
Crystal structure of mouse kynurenine aminotransferase iii, plp-bound form
|
|
Structure:
|
 |
Kynurenine-oxoglutarate transaminase 3. Chain: a, b. Synonym: kynurenine-oxoglutarate transaminase iii, kynurenine aminotransferase iii, katiii, cysteine-s-conjugate beta-lyase 2. Engineered: yes
|
|
Source:
|
 |
Mus musculus. Organism_taxid: 10090. Gene: ccbl2, kat3. Expressed in: spodoptera frugiperda. Expression_system_taxid: 7108.
|
|
Resolution:
|
 |
|
2.59Å
|
R-factor:
|
0.196
|
R-free:
|
0.246
|
|
|
Authors:
|
 |
Q.Han,R.Robinson,T.Cai,D.A.Tagle,J.Li
|
|
Key ref:
|
 |
Q.Han
et al.
(2009).
Biochemical and structural properties of mouse kynurenine aminotransferase III..
Mol cell biol,
29,
784-793.
PubMed id:
|
 |
|
Date:
|
 |
|
05-Aug-08
|
Release date:
|
30-Dec-08
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q71RI9
(KAT3_MOUSE) -
Kynurenine--oxoglutarate transaminase 3 from Mus musculus
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
455 a.a.
410 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.2.6.1.63
- kynurenine--glyoxylate transaminase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
L-kynurenine + glyoxylate = kynurenate + glycine + H2O
|
|
2.
|
3-hydroxy-L-kynurenine + glyoxylate = xanthurenate + glycine + H2O
|
|
 |
 |
 |
 |
 |
L-kynurenine
Bound ligand (Het Group name = )
matches with 57.14% similarity
|
+
|
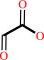 glyoxylate
glyoxylate
|
=
|
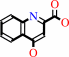 kynurenate
kynurenate
|
+
|
 glycine
glycine
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
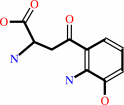 3-hydroxy-L-kynurenine
3-hydroxy-L-kynurenine
|
+
|
glyoxylate
Bound ligand (Het Group name = )
matches with 57.14% similarity
|
=
|
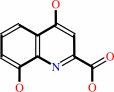 xanthurenate
xanthurenate
|
+
|
 glycine
glycine
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.2.6.1.7
- kynurenine--oxoglutarate transaminase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-kynurenine + 2-oxoglutarate = kynurenate + L-glutamate + H2O
|
 |
 |
 |
 |
 |
L-kynurenine
Bound ligand (Het Group name = )
matches with 45.45% similarity
|
+
|
 2-oxoglutarate
2-oxoglutarate
|
=
|
 kynurenate
kynurenate
|
+
|
 L-glutamate
L-glutamate
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
 Pyridoxal 5'-phosphate
Pyridoxal 5'-phosphate
|
|
 |
 |
Enzyme class 3:
|
 |
E.C.4.4.1.13
- cysteine-S-conjugate beta-lyase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
an S-substituted L-cysteine + H2O = a thiol + pyruvate + NH4+
|
 |
 |
 |
 |
 |
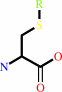 S-substituted L-cysteine
S-substituted L-cysteine
|
+
|
H2O
|
=
|
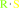 thiol
thiol
|
+
|
 pyruvate
pyruvate
|
+
|
NH4(+)
Bound ligand (Het Group name = )
matches with 71.43% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
 Pyridoxal 5'-phosphate
Pyridoxal 5'-phosphate
|
|
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

