 |
PDBsum entry 3hlm

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Crystal structure of mouse mitochondrial aspartate aminotransferase/kynurenine aminotransferase iv
|
|
Structure:
|
 |
Aspartate aminotransferase, mitochondrial. Chain: a, b, c, d. Synonym: maspat, transaminase a, glutamate oxaloacetate transaminase 2, fatty acid-binding protein, fabp-1, fabppm. Engineered: yes
|
|
Source:
|
 |
Mus musculus. Mouse. Organism_taxid: 10090. Gene: got-2, got2. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
2.50Å
|
R-factor:
|
0.184
|
R-free:
|
0.234
|
|
|
Authors:
|
 |
Q.Han,H.Robinson,J.Li
|
|
Key ref:
|
 |
Q.Han
et al.
(2010).
Structure, expression, and function of kynurenine aminotransferases in human and rodent brains.
Cell Mol Life Sci,
67,
353-368.
PubMed id:
|
 |
|
Date:
|
 |
|
27-May-09
|
Release date:
|
02-Jun-10
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P05202
(AATM_MOUSE) -
Aspartate aminotransferase, mitochondrial from Mus musculus
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
430 a.a.
401 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.2.6.1.1
- aspartate transaminase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-aspartate + 2-oxoglutarate = oxaloacetate + L-glutamate
|
 |
 |
 |
 |
 |
 L-aspartate
L-aspartate
|
+
|
 2-oxoglutarate
2-oxoglutarate
|
=
|
 oxaloacetate
oxaloacetate
|
+
|
L-glutamate
Bound ligand (Het Group name = )
matches with 50.00% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
 Pyridoxal 5'-phosphate
Pyridoxal 5'-phosphate
|
|
 |
 |
Enzyme class 2:
|
 |
E.C.2.6.1.7
- kynurenine--oxoglutarate transaminase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-kynurenine + 2-oxoglutarate = kynurenate + L-glutamate + H2O
|
 |
 |
 |
 |
 |
L-kynurenine
Bound ligand (Het Group name = )
matches with 45.45% similarity
|
+
|
 2-oxoglutarate
2-oxoglutarate
|
=
|
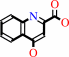 kynurenate
kynurenate
|
+
|
 L-glutamate
L-glutamate
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
 Pyridoxal 5'-phosphate
Pyridoxal 5'-phosphate
|
|
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Cell Mol Life Sci
67:353-368
(2010)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure, expression, and function of kynurenine aminotransferases in human and rodent brains.
|
|
Q.Han,
T.Cai,
D.A.Tagle,
J.Li.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Kynurenine aminotransferases (KATs) catalyze the synthesis of kynurenic acid
(KYNA), an endogenous antagonist of N-methyl-D: -aspartate and alpha 7-nicotinic
acetylcholine receptors. Abnormal KYNA levels in human brains are implicated in
the pathophysiology of schizophrenia, Alzheimer's disease, and other
neurological disorders. Four KATs have been reported in mammalian brains, KAT
I/glutamine transaminase K/cysteine conjugate beta-lyase 1, KAT II/aminoadipate
aminotransferase, KAT III/cysteine conjugate beta-lyase 2, and KAT
IV/glutamic-oxaloacetic transaminase 2/mitochondrial aspartate aminotransferase.
KAT II has a striking tertiary structure in N-terminal part and forms a new
subgroup in fold type I aminotransferases, which has been classified as subgroup
Iepsilon. Knowledge regarding KATs is vast and complex; therefore, this review
is focused on recent important progress of their gene characterization,
physiological and biochemical function, and structural properties. The
biochemical differences of four KATs, specific enzyme activity assays, and the
structural insights into the mechanism of catalysis and inhibition of these
enzymes are discussed.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
D.Zádori,
P.Klivényi,
I.Plangár,
J.Toldi,
and
L.Vécsei
(2011).
Endogenous neuroprotection in chronic neurodegenerative disorders: with particular regard to the kynurenines.
|
| |
J Cell Mol Med,
15,
701-717.
|
 |
|
|
|
|
 |
E.Passera,
B.Campanini,
F.Rossi,
V.Casazza,
M.Rizzi,
R.Pellicciari,
and
A.Mozzarelli
(2011).
Human kynurenine aminotransferase II - reactivity with substrates and inhibitors.
|
| |
FEBS J,
278,
1882-1900.
|
 |
|
|
|
|
 |
G.F.Oxenkrug
(2011).
Interferon-gamma-inducible kynurenines/pteridines inflammation cascade: implications for aging and aging-associated psychiatric and medical disorders.
|
| |
J Neural Transm,
118,
75-85.
|
 |
|
|
|
|
 |
Q.Han,
H.Robinson,
T.Cai,
D.A.Tagle,
and
J.Li
(2011).
Biochemical and structural characterization of mouse mitochondrial aspartate aminotransferase, a newly identified kynurenine aminotransferase-IV.
|
| |
Biosci Rep,
31,
323-332.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
P.Mehere,
Q.Han,
J.A.Lemkul,
C.J.Vavricka,
H.Robinson,
D.R.Bevan,
and
J.Li
(2010).
Tyrosine aminotransferase: biochemical and structural properties and molecular dynamics simulations.
|
| |
Protein Cell,
1,
1023-1032.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Q.Han,
T.Cai,
D.A.Tagle,
and
J.Li
(2010).
Thermal stability, pH dependence and inhibition of four murine kynurenine aminotransferases.
|
| |
BMC Biochem,
11,
19.
|
 |
|
|
|
|
 |
Z.T.Kincses,
J.Toldi,
and
L.Vécsei
(2010).
Kynurenines, neurodegeneration and Alzheimer's disease.
|
| |
J Cell Mol Med,
14,
2045-2054.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

