 |
PDBsum entry 1jdb

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
|
References listed in PDB file
|
 |
|
Key reference
|
 |
|
Title
|
 |
The structure of carbamoyl phosphate synthetase determined to 2.1 a resolution.
|
 |
|
Authors
|
 |
J.B.Thoden,
F.M.Raushel,
M.M.Benning,
I.Rayment,
H.M.Holden.
|
 |
|
Ref.
|
 |
Acta Crystallogr D Biol Crystallogr, 1999,
55,
8.
[DOI no: ]
|
 |
|
PubMed id
|
 |
|
 |
|
Note In the PDB file this reference is
annotated as "TO BE PUBLISHED".
The citation details given above were identified by an automated
search of PubMed on title and author
names, giving a
percentage match of
82%.
|
 |
 |
|
Abstract
|
 |
|
Carbamoyl phosphate synthetase catalyzes the formation of carbamoyl phosphate
from one molecule of bicarbonate, two molecules of Mg2+ATP and one molecule of
glutamine or ammonia depending upon the particular form of the enzyme under
investigation. As isolated from Escherichia coli, the enzyme is an
alpha,beta-heterodimer consisting of a small subunit that hydrolyzes glutamine
and a large subunit that catalyzes the two required phosphorylation events. Here
the three-dimensional structure of carbamoyl phosphate synthetase from E. coli
refined to 2.1 A resolution with an R factor of 17.9% is described. The small
subunit is distinctly bilobal with a catalytic triad (Cys269, His353 and Glu355)
situated between the two structural domains. As observed in those enzymes
belonging to the alpha/beta-hydrolase family, the active-site nucleophile,
Cys269, is perched at the top of a tight turn. The large subunit consists of
four structural units: the carboxyphosphate synthetic component, the
oligomerization domain, the carbamoyl phosphate synthetic component and the
allosteric domain. Both the carboxyphosphate and carbamoyl phosphate synthetic
components bind Mn2+ADP. In the carboxyphosphate synthetic component, the two
observed Mn2+ ions are both octahedrally coordinated by oxygen-containing
ligands and are bridged by the carboxylate side chain of Glu299. Glu215 plays a
key allosteric role by coordinating to the physiologically important potassium
ion and hydrogen bonding to the ribose hydroxyl groups of ADP. In the carbamoyl
phosphate synthetic component, the single observed Mn2+ ion is also octahedrally
coordinated by oxygen-containing ligands and Glu761 plays a similar role to that
of Glu215. The carboxyphosphate and carbamoyl phosphate synthetic components,
while topologically equivalent, are structurally different, as would be expected
in light of their separate biochemical functions.
|
 |
 |
 |

|
 |

|
 |
Figure 7.
Figure 7 The active site for the carboxyphosphate synthetic
component. A cartoon of potential hydrogen-bonding interactions
between the ADP/P[i] moiety and the protein is displayed. The
manganese ions are indicated as brown spheres.
|
 |
Figure 13.
Figure 13 A close-up view of the ornithine binding pocket.
Ordered water molecules are indicated by the red spheres. The
carboxylate group of ornithine interacts with the allosteric
domain while the 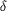 -amino
group of the side chain forms hydrogen bonds with amino-acid
residues from the carbamoyl phosphate synthetic component. -amino
group of the side chain forms hydrogen bonds with amino-acid
residues from the carbamoyl phosphate synthetic component.
|
 |
|
 |
 |
|
The above figures are
reprinted
by permission from the IUCr:
Acta Crystallogr D Biol Crystallogr
(1999,
55,
8-0)
copyright 1999.
|
 |
|
Secondary reference #1
|
 |
|
Title
|
 |
Structure of carbamoyl phosphate synthetase: a journey of 96 a from substrate to product.
|
 |
|
Authors
|
 |
J.B.Thoden,
H.M.Holden,
G.Wesenberg,
F.M.Raushel,
I.Rayment.
|
 |
|
Ref.
|
 |
Biochemistry, 1997,
36,
6305-6316.
[DOI no: ]
|
 |
|
PubMed id
|
 |
|
 |
 |
|
|
 |
|

|
|
|
 |

