 |
PDBsum entry 1e6c

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.7.1.71
- shikimate kinase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Shikimate and Chorismate Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
shikimate + ATP = 3-phosphoshikimate + ADP + H+
|
 |
 |
 |
 |
 |
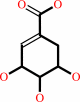 shikimate
shikimate
|
+
|
ATP
Bound ligand (Het Group name = )
matches with 66.67% similarity
|
=
|
3-phosphoshikimate
|
+
|
 ADP
ADP
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Protein Sci
10:1137-1149
(2001)
|
|
PubMed id:
|
|
|
|

|
| |
|
Biochemical and X-ray crystallographic studies on shikimate kinase: the important structural role of the P-loop lysine.
|
|
T.Krell,
J.Maclean,
D.J.Boam,
A.Cooper,
M.Resmini,
K.Brocklehurst,
S.M.Kelly,
N.C.Price,
A.J.Lapthorn,
J.R.Coggins.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Shikimate kinase, despite low sequence identity, has been shown to be
structurally a member of the nucleoside monophosphate (NMP) kinase family, which
includes adenylate kinase. In this paper we have explored the roles of residues
in the P-loop of shikimate kinase, which forms the binding site for nucleotides
and is one of the most conserved structural features in proteins. In common with
many members of the P-loop family, shikimate kinase contains a cysteine residue
2 amino acids upstream of the essential lysine residue; the side chains of these
residues are shown to form an ion pair. The C13S mutant of shikimate kinase was
found to be enzymatically active, whereas the K15M mutant was inactive. However,
the latter mutant had both increased thermostability and affinity for ATP when
compared to the wild-type enzyme. The structure of the K15M mutant protein has
been determined at 1.8 A, and shows that the organization of the P-loop and
flanking regions is heavily disturbed. This indicates that, besides its role in
catalysis, the P-loop lysine also has an important structural role. The
structure of the K15M mutant also reveals that the formation of an additional
arginine/aspartate ion pair is the most likely reason for its increased
thermostability. From studies of ligand binding it appears that, like adenylate
kinase, shikimate kinase binds substrates randomly and in a synergistic fashion,
indicating that the two enzymes have similar catalytic mechanisms.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
G.Fucile,
S.Falconer,
and
D.Christendat
(2008).
Evolutionary diversification of plant shikimate kinase gene duplicates.
|
| |
PLoS Genet,
4,
e1000292.
|
 |
|
|
|
|
 |
V.Olivares-Illana,
P.Meyer,
E.Bechet,
V.Gueguen-Chaignon,
D.Soulat,
S.Lazereg-Riquier,
I.Mijakovic,
J.Deutscher,
A.J.Cozzone,
O.Laprévote,
S.Morera,
C.Grangeasse,
and
S.Nessler
(2008).
Structural basis for the regulation mechanism of the tyrosine kinase CapB from Staphylococcus aureus.
|
| |
PLoS Biol,
6,
e143.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.V.Dias,
L.M.Faím,
I.B.Vasconcelos,
J.S.de Oliveira,
L.A.Basso,
D.S.Santos,
and
W.F.de Azevedo
(2007).
Effects of the magnesium and chloride ions and shikimate on the structure of shikimate kinase from Mycobacterium tuberculosis.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
63,
1-6.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
B.Dhaliwal,
J.Ren,
M.Lockyer,
I.Charles,
A.R.Hawkins,
and
D.K.Stammers
(2006).
Structure of Staphylococcus aureus cytidine monophosphate kinase in complex with cytidine 5'-monophosphate.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
62,
710-715.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Kotaka,
B.Dhaliwal,
J.Ren,
C.E.Nichols,
R.Angell,
M.Lockyer,
A.R.Hawkins,
and
D.K.Stammers
(2006).
Structures of S. aureus thymidylate kinase reveal an atypical active site configuration and an intermediate conformational state upon substrate binding.
|
| |
Protein Sci,
15,
774-784.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
W.C.Cheng,
Y.N.Chang,
and
W.C.Wang
(2005).
Structural basis for shikimate-binding specificity of Helicobacter pylori shikimate kinase.
|
| |
J Bacteriol,
187,
8156-8163.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.H.Pereira,
J.S.de Oliveira,
F.Canduri,
M.V.Dias,
M.S.Palma,
L.A.Basso,
D.S.Santos,
and
W.F.de Azevedo
(2004).
Structure of shikimate kinase from Mycobacterium tuberculosis reveals the binding of shikimic acid.
|
| |
Acta Crystallogr D Biol Crystallogr,
60,
2310-2319.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
E.Cerasoli,
S.M.Kelly,
J.R.Coggins,
D.J.Boam,
D.T.Clarke,
and
N.C.Price
(2002).
The refolding of type II shikimate kinase from Erwinia chrysanthemi after denaturation in urea.
|
| |
Eur J Biochem,
269,
2124-2132.
|
 |
|
|
|
|
 |
M.J.Romanowski,
and
S.K.Burley
(2002).
Crystal structure of the Escherichia coli shikimate kinase I (AroK) that confers sensitivity to mecillinam.
|
| |
Proteins,
47,
558-562.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

