
|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Structural basis for the specificity of ubiquitin c-terminal hydrolases
|
|
Structure:
|
 |
Protein (ubiquitin yuh1-ubal). Chain: a, c. Fragment: all. Engineered: yes. Other_details: ubiquitin c-terminus modified to an aldehyde. Protein (ubiquitin yuh1-ubal). Chain: b, d. Fragment: all. Engineered: yes.
|
|
Source:
|
 |
Synthetic: yes. Other_details: the protein was chemically synthesized. The sequence of this protein is naturally found in the cytoplasm of plasmid p- 1a2/trpyuh1-1 of saccharomyces cerevisiae (baker's yeast). The expression system was escherichia coli, strain mm294, plasmid p- 1a2/trpyuh1-1.. 1a2/trpyuh1-1.
|
|
Biol. unit:
|
 |
Tetramer (from
)
|
|
Resolution:
|
 |
|
2.25Å
|
R-factor:
|
0.248
|
R-free:
|
0.285
|
|
|
Authors:
|
 |
S.C.Johnston,S.M.Riddle,R.E.Cohen,C.P.Hill
|
Key ref:

|
 |
S.C.Johnston
et al.
(1999).
Structural basis for the specificity of ubiquitin C-terminal hydrolases.
EMBO J,
18,
3877-3887.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
12-May-99
|
Release date:
|
27-Jul-99
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P35127
(UBL1_YEAST) -
Ubiquitin carboxyl-terminal hydrolase YUH1 from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
236 a.a.
214 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
Chains A, C:
E.C.3.4.19.12
- ubiquitinyl hydrolase 1.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
Thiol-dependent hydrolysis of ester, thiolester, amide, peptide and isopeptide bonds formed by the C-terminal Gly of ubiquitin (a 76-residue protein attached to proteins as an intracellular targeting signal).
|
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
Chains B, D:
E.C.3.1.2.15
- Deleted entry.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
Ubiquitin C-terminal thioester + H2O = ubiquitin + a thiol
|
 |
 |
 |
 |
 |
Ubiquitin C-terminal thioester
|
+
|
H(2)O
|
=
|
ubiquitin
|
+
|
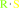 thiol
thiol
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
EMBO J
18:3877-3887
(1999)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural basis for the specificity of ubiquitin C-terminal hydrolases.
|
|
S.C.Johnston,
S.M.Riddle,
R.E.Cohen,
C.P.Hill.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The release of ubiquitin from attachment to other proteins and adducts is
critical for ubiquitin biosynthesis, proteasomal degradation and other cellular
processes. De-ubiquitination is accomplished in part by members of the UCH
(ubiquitin C-terminal hydrolase) family of enzymes. We have determined the 2.25
A resolution crystal structure of the yeast UCH, Yuh1, in a complex with the
inhibitor ubiquitin aldehyde (Ubal). The structure mimics the tetrahedral
intermediate in the reaction pathway and explains the very high enzyme
specificity. Comparison with a related, unliganded UCH structure indicates that
ubiquitin binding is coupled to rearrangements which block the active-site cleft
in the absence of authentic substrate. Remarkably, a 21-residue loop that
becomes ordered upon binding Ubal lies directly over the active site.
Efficiently processed substrates apparently pass through this loop, and
constraints on the loop conformation probably function to control UCH
specificity.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1 Stereoview ribbon representation of the Yuh1–Ubal
complex. (A) Ubal is not shown in this panel. Sidechains of the
active-site residues Gln84, Cys90, His166 and Asp181 (red) are
labeled Q, C, H and D. N- and C-termini are labeled. The
disordered segment (residues 63–77) is indicated with the
adjacent ordered residues labeled in magenta. The
active-site-crossover loop is colored yellow. Secondary
structures were as defined by PROMOTIF (Hutchinson and Thornton,
1996). Strands are colored green and the helices blue. Helix 4,
which contains the active-site nucleophile Cys90, is colored
cyan. This helix undergoes a severe kink, indeed PROMOTIF
defines residues 90–100 and 103–105 as separate helices. We
describe this as one continuous helix in order to maintain
consistency of nomenclature with UCH-L3, which also has a severe
kink in the corresponding part of helix 4. The following are
labeled on the figure: strand 0 (S0, residues 11–12), strand 1
(S1, 31–36), strand 2 (S2, 54–60), strand 2^1 (S2^1,
81–82), strand 2^2 (S2^2, 128–129), strand 3 (S3,
165–172), strand 4 (S4, 176–180), strand 5 (S5, 189–193),
strand 6 (S6, 227–233); helix 1 (H1, residues 15–25), helix
2 (H2, 44–46), helix 4 (H4, 90–105), helix 5 (H5,
111–122), helix 6 (H6, 132–143), helix 7 (H7, 205–221).
Helices are alpha-type, except for helix 2 and residues
102–105 of helix 4, which adopt the 3[10] conformation. There
is a turn of alpha helix (residues 146–150) within the
active-site-crossover loop. (B) Same as (A), but including the
Ubal shown in magenta. The sidechains of Ubal residues discussed
in the text are shown in orange and labeled. Figures 1, 4, 5 and
6 were produced with the programs MOLSCRIPT (Kraulis, 1991) and
RASTER 3D (Bacon and Anderson, 1988).
|
 |
Figure 3.
Figure 3 Schematic of the UCH/cysteine protease reaction cycle.
TI-1 and TI-2 denote the high energy tetrahedral intermediates;
AI, the acyl intermediate; S, a cysteine; and Im, a histidine.
Adapted from Storer and Ménard (1994).
|
 |
|
|

|
| |
The above figures are
reprinted
from an Open Access publication published by Macmillan Publishers Ltd:
EMBO J
(1999,
18,
3877-3887)
copyright 1999.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
C.Zheng,
Q.Yin,
and
H.Wu
(2011).
Structural studies of NF-κB signaling.
|
| |
Cell Res,
21,
183-195.
|
 |
|
|
|
|
 |
E.Kanamori,
S.Igarashi,
M.Osawa,
Y.Fukunishi,
I.Shimada,
and
H.Nakamura
(2011).
Structure determination of a protein assembly by amino acid selective cross-saturation.
|
| |
Proteins,
79,
179-190.
|
 |
|
|
|
|
 |
Y.Kodama,
M.L.Reese,
N.Shimba,
K.Ono,
E.Kanamori,
V.Dötsch,
S.Noguchi,
Y.Fukunishi,
E.Suzuki,
I.Shimada,
and
H.Takahashi
(2011).
Rapid identification of protein-protein interfaces for the construction of a complex model based on multiple unassigned signals by using time-sharing NMR measurements.
|
| |
J Struct Biol,
174,
434-442.
|
 |
|
|
|
|
 |
D.A.Boudreaux,
T.K.Maiti,
C.W.Davies,
and
C.Das
(2010).
Ubiquitin vinyl methyl ester binding orients the misaligned active site of the ubiquitin hydrolase UCHL1 into productive conformation.
|
| |
Proc Natl Acad Sci U S A,
107,
9117-9122.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.Y.Yeh,
and
P.H.Klesius
(2010).
Characterization and tissue expression of channel catfish (Ictalurus punctatus Rafinesque, 1818) ubiquitin carboxyl-terminal hydrolase L5 (UCHL5) cDNA.
|
| |
Mol Biol Rep,
37,
1229-1234.
|
 |
|
|
|
|
 |
I.N.Day,
and
R.J.Thompson
(2010).
UCHL1 (PGP 9.5): neuronal biomarker and ubiquitin system protein.
|
| |
Prog Neurobiol,
90,
327-362.
|
 |
|
|
|
|
 |
K.Artavanis-Tsakonas,
W.A.Weihofen,
J.M.Antos,
B.I.Coleman,
C.A.Comeaux,
M.T.Duraisingh,
R.Gaudet,
and
H.L.Ploegh
(2010).
Characterization and structural studies of the Plasmodium falciparum ubiquitin and Nedd8 hydrolase UCHL3.
|
| |
J Biol Chem,
285,
6857-6866.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.C.Liu,
L.Akinyi,
D.Scharf,
J.Mo,
S.F.Larner,
U.Muller,
M.W.Oli,
W.Zheng,
F.Kobeissy,
L.Papa,
X.C.Lu,
J.R.Dave,
F.C.Tortella,
R.L.Hayes,
and
K.K.Wang
(2010).
Ubiquitin C-terminal hydrolase-L1 as a biomarker for ischemic and traumatic brain injury in rats.
|
| |
Eur J Neurosci,
31,
722-732.
|
 |
|
|
|
|
 |
S.Ishii,
T.Yano,
A.Ebihara,
A.Okamoto,
M.Manzoku,
and
H.Hayashi
(2010).
Crystal structure of the peptidase domain of Streptococcus ComA, a bifunctional ATP-binding cassette transporter involved in the quorum-sensing pathway.
|
| |
J Biol Chem,
285,
10777-10785.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Ramakrishna,
B.Suresh,
I.C.Kang,
and
K.H.Baek
(2010).
Polyclonal and monoclonal antibodies specific for USP17, a proapoptotic deubiquitinating enzyme.
|
| |
Hybridoma (Larchmt),
29,
311-319.
|
 |
|
|
|
|
 |
D.Komander,
F.Reyes-Turcu,
J.D.Licchesi,
P.Odenwaelder,
K.D.Wilkinson,
and
D.Barford
(2009).
Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains.
|
| |
EMBO Rep,
10,
466-473.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.Komander,
M.J.Clague,
and
S.Urbé
(2009).
Breaking the chains: structure and function of the deubiquitinases.
|
| |
Nat Rev Mol Cell Biol,
10,
550-563.
|
 |
|
|
|
|
 |
F.E.Reyes-Turcu,
and
K.D.Wilkinson
(2009).
Polyubiquitin binding and disassembly by deubiquitinating enzymes.
|
| |
Chem Rev,
109,
1495-1508.
|
 |
|
|
|
|
 |
F.E.Reyes-Turcu,
K.H.Ventii,
and
K.D.Wilkinson
(2009).
Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes.
|
| |
Annu Rev Biochem,
78,
363-397.
|
 |
|
|
|
|
 |
F.I.Andersson,
D.G.Pina,
A.L.Mallam,
G.Blaser,
and
S.E.Jackson
(2009).
Untangling the folding mechanism of the 5-knotted protein UCH-L3.
|
| |
FEBS J,
276,
2625-2635.
|
 |
|
|
|
|
 |
F.K.Insaidoo,
J.Zajicek,
and
B.M.Baker
(2009).
A general and efficient approach for NMR studies of peptide dynamics in class I MHC peptide binding grooves.
|
| |
Biochemistry,
48,
9708-9710.
|
 |
|
|
|
|
 |
G.Nicastro,
L.Masino,
V.Esposito,
R.P.Menon,
A.De Simone,
F.Fraternali,
and
A.Pastore
(2009).
Josephin domain of ataxin-3 contains two distinct ubiquitin-binding sites.
|
| |
Biopolymers,
91,
1203-1214.
|
 |
|
|
|
|
 |
M.W.Foster,
M.T.Forrester,
and
J.S.Stamler
(2009).
A protein microarray-based analysis of S-nitrosylation.
|
| |
Proc Natl Acad Sci U S A,
106,
18948-18953.
|
 |
|
|
|
|
 |
M.W.Popp,
K.Artavanis-Tsakonas,
and
H.L.Ploegh
(2009).
Substrate Filtering by the Active Site Crossover Loop in UCHL3 Revealed by Sortagging and Gain-of-function Mutations.
|
| |
J Biol Chem,
284,
3593-3602.
|
 |
|
|
|
|
 |
T.S.Kroeger,
K.P.Watkins,
G.Friso,
K.J.van Wijk,
and
A.Barkan
(2009).
A plant-specific RNA-binding domain revealed through analysis of chloroplast group II intron splicing.
|
| |
Proc Natl Acad Sci U S A,
106,
4537-4542.
|
 |
|
|
|
|
 |
A.M.Burroughs,
M.Jaffee,
L.M.Iyer,
and
L.Aravind
(2008).
Anatomy of the E2 ligase fold: implications for enzymology and evolution of ubiquitin/Ub-like protein conjugation.
|
| |
J Struct Biol,
162,
205-218.
|
 |
|
|
|
|
 |
G.Rabut,
and
M.Peter
(2008).
Function and regulation of protein neddylation. 'Protein modifications: beyond the usual suspects' review series.
|
| |
EMBO Rep,
9,
969-976.
|
 |
|
|
|
|
 |
J.Souphron,
M.B.Waddell,
A.Paydar,
Z.Tokgöz-Gromley,
M.F.Roussel,
and
B.A.Schulman
(2008).
Structural dissection of a gating mechanism preventing misactivation of ubiquitin by NEDD8's E1.
|
| |
Biochemistry,
47,
8961-8969.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
K.H.Ventii,
and
K.D.Wilkinson
(2008).
Protein partners of deubiquitinating enzymes.
|
| |
Biochem J,
414,
161-175.
|
 |
|
|
|
|
 |
L.Song,
and
M.Rape
(2008).
Reverse the curse--the role of deubiquitination in cell cycle control.
|
| |
Curr Opin Cell Biol,
20,
156-163.
|
 |
|
|
|
|
 |
N.A.Lakomek,
K.F.Walter,
C.Farès,
O.F.Lange,
B.L.de Groot,
H.Grubmüller,
R.Brüschweiler,
A.Munk,
S.Becker,
J.Meiler,
and
C.Griesinger
(2008).
Self-consistent residual dipolar coupling based model-free analysis for the robust determination of nanosecond to microsecond protein dynamics.
|
| |
J Biomol NMR,
41,
139-155.
|
 |
|
|
|
|
 |
O.Riess,
U.Rüb,
A.Pastore,
P.Bauer,
and
L.Schöls
(2008).
SCA3: Neurological features, pathogenesis and animal models.
|
| |
Cerebellum,
7,
125-137.
|
 |
|
|
|
|
 |
S.C.Lin,
J.Y.Chung,
B.Lamothe,
K.Rajashankar,
M.Lu,
Y.C.Lo,
A.Y.Lam,
B.G.Darnay,
and
H.Wu
(2008).
Molecular basis for the unique deubiquitinating activity of the NF-kappaB inhibitor A20.
|
| |
J Mol Biol,
376,
526-540.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.Kabuta,
A.Furuta,
S.Aoki,
K.Furuta,
and
K.Wada
(2008).
Aberrant Interaction between Parkinson Disease-associated Mutant UCH-L1 and the Lysosomal Receptor for Chaperone-mediated Autophagy.
|
| |
J Biol Chem,
283,
23731-23738.
|
 |
|
|
|
|
 |
T.Yao,
L.Song,
J.Jin,
Y.Cai,
H.Takahashi,
S.K.Swanson,
M.P.Washburn,
L.Florens,
R.C.Conaway,
R.E.Cohen,
and
J.W.Conaway
(2008).
Distinct modes of regulation of the Uch37 deubiquitinating enzyme in the proteasome and in the Ino80 chromatin-remodeling complex.
|
| |
Mol Cell,
31,
909-917.
|
 |
|
|
|
|
 |
Y.Liu,
F.Wang,
H.Zhang,
H.He,
L.Ma,
and
X.W.Deng
(2008).
Functional characterization of the Arabidopsis ubiquitin-specific protease gene family reveals specific role and redundancy of individual members in development.
|
| |
Plant J,
55,
844-856.
|
 |
|
|
|
|
 |
A.D.Capili,
and
C.D.Lima
(2007).
Taking it step by step: mechanistic insights from structural studies of ubiquitin/ubiquitin-like protein modification pathways.
|
| |
Curr Opin Struct Biol,
17,
726-735.
|
 |
|
|
|
|
 |
A.Fernández-Montalván,
T.Bouwmeester,
G.Joberty,
R.Mader,
M.Mahnke,
B.Pierrat,
J.M.Schlaeppi,
S.Worpenberg,
and
B.Gerhartz
(2007).
Biochemical characterization of USP7 reveals post-translational modification sites and structural requirements for substrate processing and subcellular localization.
|
| |
FEBS J,
274,
4256-4270.
|
 |
|
|
|
|
 |
C.Richter,
M.West,
and
G.Odorizzi
(2007).
Dual mechanisms specify Doa4-mediated deubiquitination at multivesicular bodies.
|
| |
EMBO J,
26,
2454-2464.
|
 |
|
|
|
|
 |
C.Schlieker,
W.A.Weihofen,
E.Frijns,
L.M.Kattenhorn,
R.Gaudet,
and
H.L.Ploegh
(2007).
Structure of a herpesvirus-encoded cysteine protease reveals a unique class of deubiquitinating enzymes.
|
| |
Mol Cell,
25,
677-687.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
E.M.Frickel,
V.Quesada,
L.Muething,
M.J.Gubbels,
E.Spooner,
H.Ploegh,
and
K.Artavanis-Tsakonas
(2007).
Apicomplexan UCHL3 retains dual specificity for ubiquitin and Nedd8 throughout evolution.
|
| |
Cell Microbiol,
9,
1601-1610.
|
 |
|
|
|
|
 |
R.K.Meray,
and
P.T.Lansbury
(2007).
Reversible monoubiquitination regulates the Parkinson disease-associated ubiquitin hydrolase UCH-L1.
|
| |
J Biol Chem,
282,
10567-10575.
|
 |
|
|
|
|
 |
B.M.Kessler
(2006).
Putting proteomics on target: activity-based profiling of ubiquitin and ubiquitin-like processing enzymes.
|
| |
Expert Rev Proteomics,
3,
213-221.
|
 |
|
|
|
|
 |
C.Das,
Q.Q.Hoang,
C.A.Kreinbring,
S.J.Luchansky,
R.K.Meray,
S.S.Ray,
P.T.Lansbury,
D.Ringe,
and
G.A.Petsko
(2006).
Structural basis for conformational plasticity of the Parkinson's disease-associated ubiquitin hydrolase UCH-L1.
|
| |
Proc Natl Acad Sci U S A,
103,
4675-4680.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.Reverter,
and
C.D.Lima
(2006).
Structural basis for SENP2 protease interactions with SUMO precursors and conjugated substrates.
|
| |
Nat Struct Mol Biol,
13,
1060-1068.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.T.Huang,
and
B.A.Schulman
(2006).
Breaking up with a kinky SUMO.
|
| |
Nat Struct Mol Biol,
13,
1045-1047.
|
 |
|
|
|
|
 |
G.Nicastro,
M.Habeck,
L.Masino,
D.I.Svergun,
and
A.Pastore
(2006).
Structure validation of the Josephin domain of ataxin-3: conclusive evidence for an open conformation.
|
| |
J Biomol NMR,
36,
267-277.
|
 |
|
|
|
|
 |
J.J.Arnold,
A.Bernal,
U.Uche,
D.E.Sterner,
T.R.Butt,
C.E.Cameron,
and
M.R.Mattern
(2006).
Small ubiquitin-like modifying protein isopeptidase assay based on poliovirus RNA polymerase activity.
|
| |
Anal Biochem,
350,
214-221.
|
 |
|
|
|
|
 |
L.Shen,
M.H.Tatham,
C.Dong,
A.Zagórska,
J.H.Naismith,
and
R.T.Hay
(2006).
SUMO protease SENP1 induces isomerization of the scissile peptide bond.
|
| |
Nat Struct Mol Biol,
13,
1069-1077.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
P.Virnau,
L.A.Mirny,
and
M.Kardar
(2006).
Intricate knots in proteins: Function and evolution.
|
| |
PLoS Comput Biol,
2,
e122.
|
 |
|
|
|
|
 |
S.Ishii,
T.Yano,
and
H.Hayashi
(2006).
Expression and characterization of the peptidase domain of Streptococcus pneumoniae ComA, a bifunctional ATP-binding cassette transporter involved in quorum sensing pathway.
|
| |
J Biol Chem,
281,
4726-4731.
|
 |
|
|
|
|
 |
T.Sakai,
H.Tochio,
T.Tenno,
Y.Ito,
T.Kokubo,
H.Hiroaki,
and
M.Shirakawa
(2006).
In-cell NMR spectroscopy of proteins inside Xenopus laevis oocytes.
|
| |
J Biomol NMR,
36,
179-188.
|
 |
|
|
|
|
 |
T.Sulea,
H.A.Lindner,
and
R.Ménard
(2006).
Structural aspects of recently discovered viral deubiquitinating activities.
|
| |
Biol Chem,
387,
853-862.
|
 |
|
|
|
|
 |
T.Yao,
L.Song,
W.Xu,
G.N.DeMartino,
L.Florens,
S.K.Swanson,
M.P.Washburn,
R.C.Conaway,
J.W.Conaway,
and
R.E.Cohen
(2006).
Proteasome recruitment and activation of the Uch37 deubiquitinating enzyme by Adrm1.
|
| |
Nat Cell Biol,
8,
994.
|
 |
|
|
|
|
 |
G.Nicastro,
R.P.Menon,
L.Masino,
P.P.Knowles,
N.Q.McDonald,
and
A.Pastore
(2005).
The solution structure of the Josephin domain of ataxin-3: structural determinants for molecular recognition.
|
| |
Proc Natl Acad Sci U S A,
102,
10493-10498.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
H.A.Lindner,
N.Fotouhi-Ardakani,
V.Lytvyn,
P.Lachance,
T.Sulea,
and
R.Ménard
(2005).
The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme.
|
| |
J Virol,
79,
15199-15208.
|
 |
|
|
|
|
 |
I.A.Rose
(2005).
Ubiquitin at Fox Chase.
|
| |
Proc Natl Acad Sci U S A,
102,
11575-11577.
|
 |
|
|
|
|
 |
I.Rose
(2005).
Ubiquitin at Fox Chase.
|
| |
Cell Death Differ,
12,
1198-1201.
|
 |
|
|
|
|
 |
I.Rose
(2005).
Ubiquitin at Fox Chase (Nobel lecture).
|
| |
Angew Chem Int Ed Engl,
44,
5926-5931.
|
 |
|
|
|
|
 |
K.Sugawara,
N.N.Suzuki,
Y.Fujioka,
N.Mizushima,
Y.Ohsumi,
and
F.Inagaki
(2005).
Structural basis for the specificity and catalysis of human Atg4B responsible for mammalian autophagy.
|
| |
J Biol Chem,
280,
40058-40065.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
L.Hicke,
H.L.Schubert,
and
C.P.Hill
(2005).
Ubiquitin-binding domains.
|
| |
Nat Rev Mol Cell Biol,
6,
610-621.
|
 |
|
|
|
|
 |
L.N.Shen,
H.Liu,
C.Dong,
D.Xirodimas,
J.H.Naismith,
and
R.T.Hay
(2005).
Structural basis of NEDD8 ubiquitin discrimination by the deNEDDylating enzyme NEDP1.
|
| |
EMBO J,
24,
1341-1351.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Hu,
P.Li,
L.Song,
P.D.Jeffrey,
T.A.Chenova,
K.D.Wilkinson,
R.E.Cohen,
and
Y.Shi
(2005).
Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14.
|
| |
EMBO J,
24,
3747-3756.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
N.Barretto,
D.Jukneliene,
K.Ratia,
Z.Chen,
A.D.Mesecar,
and
S.C.Baker
(2005).
The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity.
|
| |
J Virol,
79,
15189-15198.
|
 |
|
|
|
|
 |
S.Misaghi,
P.J.Galardy,
W.J.Meester,
H.Ovaa,
H.L.Ploegh,
and
R.Gaudet
(2005).
Structure of the ubiquitin hydrolase UCH-L3 complexed with a suicide substrate.
|
| |
J Biol Chem,
280,
1512-1520.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Y.Mao,
F.Senic-Matuglia,
P.P.Di Fiore,
S.Polo,
M.E.Hodsdon,
and
P.De Camilli
(2005).
Deubiquitinating function of ataxin-3: insights from the solution structure of the Josephin domain.
|
| |
Proc Natl Acad Sci U S A,
102,
12700-12705.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Guterman,
and
M.H.Glickman
(2004).
Complementary roles for Rpn11 and Ubp6 in deubiquitination and proteolysis by the proteasome.
|
| |
J Biol Chem,
279,
1729-1738.
|
 |
|
|
|
|
 |
A.M.Catanzariti,
T.A.Soboleva,
D.A.Jans,
P.G.Board,
and
R.T.Baker
(2004).
An efficient system for high-level expression and easy purification of authentic recombinant proteins.
|
| |
Protein Sci,
13,
1331-1339.
|
 |
|
|
|
|
 |
D.Reverter,
and
C.D.Lima
(2004).
A basis for SUMO protease specificity provided by analysis of human Senp2 and a Senp2-SUMO complex.
|
| |
Structure,
12,
1519-1531.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.T.Huang,
D.W.Miller,
R.Mathew,
R.Cassell,
J.M.Holton,
M.F.Roussel,
and
B.A.Schulman
(2004).
A unique E1-E2 interaction required for optimal conjugation of the ubiquitin-like protein NEDD8.
|
| |
Nat Struct Mol Biol,
11,
927-935.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.Hemelaar,
A.Borodovsky,
B.M.Kessler,
D.Reverter,
J.Cook,
N.Kolli,
T.Gan-Erdene,
K.D.Wilkinson,
G.Gill,
C.D.Lima,
H.L.Ploegh,
and
H.Ovaa
(2004).
Specific and covalent targeting of conjugating and deconjugating enzymes of ubiquitin-like proteins.
|
| |
Mol Cell Biol,
24,
84-95.
|
 |
|
|
|
|
 |
M.Albrecht,
M.Golatta,
U.Wüllner,
and
T.Lengauer
(2004).
Structural and functional analysis of ataxin-2 and ataxin-3.
|
| |
Eur J Biochem,
271,
3155-3170.
|
 |
|
|
|
|
 |
M.H.Nanao,
S.O.Tcherniuk,
J.Chroboczek,
O.Dideberg,
A.Dessen,
and
M.Y.Balakirev
(2004).
Crystal structure of human otubain 2.
|
| |
EMBO Rep,
5,
783-788.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
X.I.Ambroggio,
D.C.Rees,
and
R.J.Deshaies
(2004).
JAMM: a metalloprotease-like zinc site in the proteasome and signalosome.
|
| |
PLoS Biol,
2,
E2.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.P.VanDemark,
and
C.P.Hill
(2003).
Two-stepping with E1.
|
| |
Nat Struct Biol,
10,
244-246.
|
 |
|
|
|
|
 |
B.R.Wong,
F.Parlati,
K.Qu,
S.Demo,
T.Pray,
J.Huang,
D.G.Payan,
and
M.K.Bennett
(2003).
Drug discovery in the ubiquitin regulatory pathway.
|
| |
Drug Discov Today,
8,
746-754.
|
 |
|
|
|
|
 |
C.D.Lima
(2003).
Regulating UBP-mediated ubiquitin deconjugation.
|
| |
Structure,
11,
3-4.
|
 |
|
|
|
|
 |
J.Hemelaar,
V.S.Lelyveld,
B.M.Kessler,
and
H.L.Ploegh
(2003).
A single protease, Apg4B, is specific for the autophagy-related ubiquitin-like proteins GATE-16, MAP1-LC3, GABARAP, and Apg8L.
|
| |
J Biol Chem,
278,
51841-51850.
|
 |
|
|
|
|
 |
M.Sulpizi,
A.Laio,
J.VandeVondele,
A.Cattaneo,
U.Rothlisberger,
and
P.Carloni
(2003).
Reaction mechanism of caspases: insights from QM/MM Car-Parrinello simulations.
|
| |
Proteins,
52,
212-224.
|
 |
|
|
|
|
 |
M.Sulpizi,
U.Rothlisberger,
and
P.Carloni
(2003).
Molecular dynamics studies of caspase-3.
|
| |
Biophys J,
84,
2207-2215.
|
 |
|
|
|
|
 |
M.Y.Balakirev,
S.O.Tcherniuk,
M.Jaquinod,
and
J.Chroboczek
(2003).
Otubains: a new family of cysteine proteases in the ubiquitin pathway.
|
| |
EMBO Rep,
4,
517-522.
|
 |
|
|
|
|
 |
P.Y.Wu,
M.Hanlon,
M.Eddins,
C.Tsui,
R.S.Rogers,
J.P.Jensen,
M.J.Matunis,
A.M.Weissman,
A.M.Weisman,
A.M.Weissman,
C.Wolberger,
C.P.Wolberger,
and
C.M.Pickart
(2003).
A conserved catalytic residue in the ubiquitin-conjugating enzyme family.
|
| |
EMBO J,
22,
5241-5250.
|
 |
|
|
|
|
 |
S.S.Wing
(2003).
Deubiquitinating enzymes--the importance of driving in reverse along the ubiquitin-proteasome pathway.
|
| |
Int J Biochem Cell Biol,
35,
590-605.
|
 |
|
|
|
|
 |
T.Gan-Erdene,
K.Nagamalleswari,
L.Yin,
K.Wu,
Z.Q.Pan,
and
K.D.Wilkinson
(2003).
Identification and characterization of DEN1, a deneddylase of the ULP family.
|
| |
J Biol Chem,
278,
28892-28900.
|
 |
|
|
|
|
 |
Y.Liu,
H.A.Lashuel,
S.Choi,
X.Xing,
A.Case,
J.Ni,
L.A.Yeh,
G.D.Cuny,
R.L.Stein,
and
P.T.Lansbury
(2003).
Discovery of inhibitors that elucidate the role of UCH-L1 activity in the H1299 lung cancer cell line.
|
| |
Chem Biol,
10,
837-846.
|
 |
|
|
|
|
 |
M.Hu,
P.Li,
M.Li,
W.Li,
T.Yao,
J.W.Wu,
W.Gu,
R.E.Cohen,
and
Y.Shi
(2002).
Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde.
|
| |
Cell,
111,
1041-1054.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Y.Balakirev,
M.Jaquinod,
A.L.Haas,
and
J.Chroboczek
(2002).
Deubiquitinating function of adenovirus proteinase.
|
| |
J Virol,
76,
6323-6331.
|
 |
|
|
|
|
 |
Y.Liu,
L.Fallon,
H.A.Lashuel,
Z.Liu,
and
P.T.Lansbury
(2002).
The UCH-L1 gene encodes two opposing enzymatic activities that affect alpha-synuclein degradation and Parkinson's disease susceptibility.
|
| |
Cell,
111,
209-218.
|
 |
|
|
|
|
 |
C.Ingvardsen,
and
B.Veierskov
(2001).
Ubiquitin- and proteasome-dependent proteolysis in plants.
|
| |
Physiol Plant,
112,
451-459.
|
 |
|
|
|
|
 |
C.M.Pickart
(2001).
Mechanisms underlying ubiquitination.
|
| |
Annu Rev Biochem,
70,
503-533.
|
 |
|
|
|
|
 |
E.Mossessova,
and
C.D.Lima
(2000).
Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast.
|
| |
Mol Cell,
5,
865-876.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
H.Lin,
A.Keriel,
C.R.Morales,
N.Bedard,
Q.Zhao,
P.Hingamp,
S.Lefrançois,
L.Combaret,
and
S.S.Wing
(2000).
Divergent N-terminal sequences target an inducible testis deubiquitinating enzyme to distinct subcellular structures.
|
| |
Mol Cell Biol,
20,
6568-6578.
|
 |
|
|
|
|
 |
K.D.Wilkinson
(2000).
Ubiquitination and deubiquitination: targeting of proteins for degradation by the proteasome.
|
| |
Semin Cell Dev Biol,
11,
141-148.
|
 |
|
|
|
|
 |
L.Huang,
E.Kinnucan,
G.Wang,
S.Beaudenon,
P.M.Howley,
J.M.Huibregtse,
and
N.P.Pavletich
(1999).
Structure of an E6AP-UbcH7 complex: insights into ubiquitination by the E2-E3 enzyme cascade.
|
| |
Science,
286,
1321-1326.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |
|

