 |
PDBsum entry 3dba

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Crystal structure of the cgmp-bound gaf a domain from the photoreceptor phosphodiesterase 6c
|
|
Structure:
|
 |
Cone cgmp-specific 3',5'-cyclic phosphodiesterase subunit alpha'. Chain: a, b. Fragment: gaf a domain (unp residues 55-225). Synonym: cgmp phosphodiesterase 6c. Engineered: yes
|
|
Source:
|
 |
Gallus gallus. Bantam,chickens. Organism_taxid: 9031. Strain: rhode island red. Gene: pde6c. Expressed in: escherichia coli.
|
|
Resolution:
|
 |
|
2.57Å
|
R-factor:
|
0.222
|
R-free:
|
0.257
|
|
|
Authors:
|
 |
S.E.Martinez,C.C.Heikaus,R.E.Klevit,J.A.Beavo
|
|
Key ref:
|
 |
S.E.Martinez
et al.
(2008).
The structure of the GAF A domain from phosphodiesterase 6C reveals determinants of cGMP binding, a conserved binding surface, and a large cGMP-dependent conformational change.
J Biol Chem,
283,
25913-25919.
PubMed id:
|
 |
|
Date:
|
 |
|
30-May-08
|
Release date:
|
01-Jul-08
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P52731
(PDE6C_CHICK) -
Cone cGMP-specific 3',5'-cyclic phosphodiesterase subunit alpha' from Gallus gallus
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
862 a.a.
171 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.1.4.35
- 3',5'-cyclic-GMP phosphodiesterase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
3',5'-cyclic GMP + H2O = GMP + H+
|
 |
 |
 |
 |
 |
3',5'-cyclic GMP
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
H2O
|
=
|
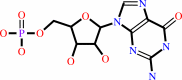 GMP
GMP
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
J Biol Chem
283:25913-25919
(2008)
|
|
PubMed id:
|
|
|
|

|
| |
|
The structure of the GAF A domain from phosphodiesterase 6C reveals determinants of cGMP binding, a conserved binding surface, and a large cGMP-dependent conformational change.
|
|
S.E.Martinez,
C.C.Heikaus,
R.E.Klevit,
J.A.Beavo.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The photoreceptor phosphodiesterase (PDE6) regulates the intracellular levels of
the second messenger cGMP in the outer segments of cone and rod photoreceptor
cells. PDE6 contains two regulatory GAF domains, of which one (GAF A) binds cGMP
and regulates the activity of the PDE6 holoenzyme. To increase our understanding
of this allosteric regulation mechanism, we present the 2.6A crystal structure
of the cGMP-bound GAF A domain of chicken cone PDE6. Nucleotide specificity
appears to be provided in part by the orientation of Asn-116, which makes two
hydrogen bonds to the guanine ring of cGMP but is not strictly conserved among
PDE6 isoforms. The isolated PDE6C GAF A domain is monomeric and does not contain
sufficient structural determinants to form a homodimer as found in full-length
PDE6C. A highly conserved surface patch on GAF A indicates a potential binding
site for the inhibitory subunit Pgamma. NMR studies reveal that the apo-PDE6C
GAF A domain is structured but adopts a significantly altered structural state
indicating a large conformational change with rearrangement of secondary
structure elements upon cGMP binding. The presented crystal structure will help
to define the cGMP-dependent regulation mechanism of the PDE6 holoenzyme and its
inhibition through Pgamma binding.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.Yamazaki,
F.Hayashi,
I.Matsuura,
and
V.A.Bondarenko
(2011).
Binding of cGMP to the transducin-activated cGMP phosphodiesterase, PDE6, initiates a large conformational change involved in its deactivation.
|
| |
FEBS J,
278,
1854-1872.
|
 |
|
|
|
|
 |
M.Russwurm,
C.Schlicker,
M.Weyand,
D.Koesling,
and
C.Steegborn
(2011).
Crystal structure of the GAF-B domain from human phosphodiesterase 5.
|
| |
Proteins,
79,
1682-1687.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.Grau,
N.O.Artemyev,
T.Rosenberg,
H.Dollfus,
O.H.Haugen,
E.Cumhur Sener,
B.Jurklies,
S.Andreasson,
C.Kernstock,
M.Larsen,
E.Zrenner,
B.Wissinger,
and
S.Kohl
(2011).
Decreased catalytic activity and altered activation properties of PDE6C mutants associated with autosomal recessive achromatopsia.
|
| |
Hum Mol Genet,
20,
719-730.
|
 |
|
|
|
|
 |
A.A.Thiadens,
A.I.den Hollander,
S.Roosing,
S.B.Nabuurs,
R.C.Zekveld-Vroon,
R.W.Collin,
E.De Baere,
R.K.Koenekoop,
M.J.van Schooneveld,
T.M.Strom,
J.J.van Lith-Verhoeven,
A.J.Lotery,
N.van Moll-Ramirez,
B.P.Leroy,
L.I.van den Born,
C.B.Hoyng,
F.P.Cremers,
and
C.C.Klaver
(2009).
Homozygosity mapping reveals PDE6C mutations in patients with early-onset cone photoreceptor disorders.
|
| |
Am J Hum Genet,
85,
240-247.
|
 |
|
|
|
|
 |
C.C.Heikaus,
J.Pandit,
and
R.E.Klevit
(2009).
Cyclic nucleotide binding GAF domains from phosphodiesterases: structural and mechanistic insights.
|
| |
Structure,
17,
1551-1557.
|
 |
|
|
|
|
 |
J.Pandit,
M.D.Forman,
K.F.Fennell,
K.S.Dillman,
and
F.S.Menniti
(2009).
Mechanism for the allosteric regulation of phosphodiesterase 2A deduced from the X-ray structure of a near full-length construct.
|
| |
Proc Natl Acad Sci U S A,
106,
18225-18230.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.T.Liu,
S.L.Matte,
J.D.Corbin,
S.H.Francis,
and
R.H.Cote
(2009).
Probing the catalytic sites and activation mechanism of photoreceptor phosphodiesterase using radiolabeled phosphodiesterase inhibitors.
|
| |
J Biol Chem,
284,
31541-31547.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

