 |
PDBsum entry 1map

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Aminotransferase
|
PDB id
|
|

|

|
1map
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.2.6.1.1
- aspartate transaminase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-aspartate + 2-oxoglutarate = oxaloacetate + L-glutamate
|
 |
 |
 |
 |
 |
 L-aspartate
L-aspartate
|
+
|
 2-oxoglutarate
2-oxoglutarate
|
=
|
 oxaloacetate
oxaloacetate
|
+
|
 L-glutamate
L-glutamate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
KET)
matches with 60.00% similarity
|
|
 |
 |
Enzyme class 2:
|
 |
E.C.2.6.1.7
- kynurenine--oxoglutarate transaminase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-kynurenine + 2-oxoglutarate = kynurenate + L-glutamate + H2O
|
 |
 |
 |
 |
 |
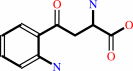 L-kynurenine
L-kynurenine
|
+
|
 2-oxoglutarate
2-oxoglutarate
|
=
|
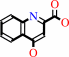 kynurenate
kynurenate
|
+
|
 L-glutamate
L-glutamate
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
KET)
matches with 60.00% similarity
|
|
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
32:13451-13462
(1993)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structures of true enzymatic reaction intermediates: aspartate and glutamate ketimines in aspartate aminotransferase.
|
|
V.N.Malashkevich,
M.D.Toney,
J.N.Jansonius.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The crystal structures of the stable, closed complexes of chicken mitochondrial
aspartate aminotransferase with the natural substrates L-aspartate and
L-glutamate have been solved and refined at 2.4- and 2.3-A resolution,
respectively. In both cases, clear electron density at the substrate-coenzyme
binding site unequivocally indicates the presence of a covalent intermediate.
The crystallographically identical environments of the two subunits of the alpha
2 dimer allow a simple, direct correlation of the coenzyme absorption spectra of
the crystalline enzyme with the diffraction results. Deconvolution of the
spectra of the crystalline complexes using lognormal curves indicates that the
ketimine intermediates constitute 76% and 83% of the total enzyme populations
with L-aspartate and L-glutamate, respectively. The electron density maps
accommodate the ketimine structures best in agreement with the independent
spectral data. Crystalline enzyme has a much higher affinity for keto acid
substrates compared to enzyme in solution. The increased affinity is interpreted
in terms of a perturbation of the open/closed conformational equilibrium by the
crystal lattice, with the closed form having greater affinity for substrate. The
crystal lattice contacts provide energy required for domain closure normally
supplied by the excess binding energy of the substrate. In solution, enzyme
saturated with amino/keto acid substrate pairs has a greater total fraction of
intermediates in the aldehyde oxidation state compared to crystalline enzyme.
Assuming the only difference between the solution and crystalline enzymes is in
conformational freedom, this difference suggests that one or more substantially
populated, aldehydic intermediates in solution exist in the open conformation.
Quantitative analyses of the spectra indicate that the value of the equilibrium
constant for the open-closed conformational transition of the liganded,
aldehydic enzyme in solution is near 1. The C4' pro-S proton in the ketimine
models is oriented nearly perpendicularly to the plane of the pyridine ring,
suggesting that the enzyme facilitates its removal by maximizing sigma-pi
orbital overlap. The absence of a localized water molecule near Lys258 dictates
that ketimine hydrolysis occurs via a transiently bound water molecule or from
an alternative, possibly more open, structure in which water is appropriately
bound. A prominent mechanistic role for flexibility of the Lys258 side chain is
suggested by the absence of hydrogen bonds to the amino group in the aspartate
structure and the relatively high temperature factors for these atoms in both
structures.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
P.D.Cook,
and
H.M.Holden
(2007).
A structural study of GDP-4-keto-6-deoxy-D-mannose-3-dehydratase: caught in the act of geminal diamine formation.
|
| |
Biochemistry,
46,
14215-14224.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
B.Golinelli-Pimpaneau,
C.Lüthi,
and
P.Christen
(2006).
Structural basis for D-amino acid transamination by the pyridoxal 5'-phosphate-dependent catalytic antibody 15A9.
|
| |
J Biol Chem,
281,
23969-23977.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
P.D.Cook,
J.B.Thoden,
and
H.M.Holden
(2006).
The structure of GDP-4-keto-6-deoxy-D-mannose-3-dehydratase: a unique coenzyme B6-dependent enzyme.
|
| |
Protein Sci,
15,
2093-2106.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
B.Adams,
K.Lowpetch,
F.Thorndycroft,
S.M.Whyte,
and
D.W.Young
(2005).
Stereochemistry of reactions of the inhibitor/substrates L- and D-beta-chloroalanine with beta-mercaptoethanol catalysed by L-aspartate aminotransferase and D-amino acid aminotransferase respectively.
|
| |
Org Biomol Chem,
3,
3357-3364.
|
 |
|
|
|
|
 |
K.Hirotsu,
M.Goto,
A.Okamoto,
and
I.Miyahara
(2005).
Dual substrate recognition of aminotransferases.
|
| |
Chem Rec,
5,
160-172.
|
 |
|
|
|
|
 |
B.Pioselli,
S.Bettati,
T.V.Demidkina,
L.N.Zakomirdina,
R.S.Phillips,
and
A.Mozzarelli
(2004).
Tyrosine phenol-lyase and tryptophan indole-lyase encapsulated in wet nanoporous silica gels: Selective stabilization of tertiary conformations.
|
| |
Protein Sci,
13,
913-924.
|
 |
|
|
|
|
 |
K.Das,
G.H.Butler,
V.Kwiatkowski,
A.D.Clark,
P.Yadav,
and
E.Arnold
(2004).
Crystal structures of arginine deiminase with covalent reaction intermediates; implications for catalytic mechanism.
|
| |
Structure,
12,
657-667.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.Schwarzenbacher,
L.Jaroszewski,
F.von Delft,
P.Abdubek,
E.Ambing,
T.Biorac,
L.S.Brinen,
J.M.Canaves,
J.Cambell,
H.J.Chiu,
X.Dai,
A.M.Deacon,
M.DiDonato,
M.A.Elsliger,
S.Eshagi,
R.Floyd,
A.Godzik,
C.Grittini,
S.K.Grzechnik,
E.Hampton,
C.Karlak,
H.E.Klock,
E.Koesema,
J.S.Kovarik,
A.Kreusch,
P.Kuhn,
S.A.Lesley,
I.Levin,
D.McMullan,
T.M.McPhillips,
M.D.Miller,
A.Morse,
K.Moy,
J.Ouyang,
R.Page,
K.Quijano,
A.Robb,
G.Spraggon,
R.C.Stevens,
H.van den Bedem,
J.Velasquez,
J.Vincent,
X.Wang,
B.West,
G.Wolf,
Q.Xu,
K.O.Hodgson,
J.Wooley,
and
I.A.Wilson
(2004).
Crystal structure of an aspartate aminotransferase (TM1255) from Thermotoga maritima at 1.90 A resolution.
|
| |
Proteins,
55,
759-763.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.C.Rothman,
M.Voorhies,
and
J.F.Kirsch
(2004).
Directed evolution relieves product inhibition and confers in vivo function to a rationally designed tyrosine aminotransferase.
|
| |
Protein Sci,
13,
763-772.
|
 |
|
|
|
|
 |
H.Hayashi,
H.Mizuguchi,
I.Miyahara,
Y.Nakajima,
K.Hirotsu,
and
H.Kagamiyama
(2003).
Conformational change in aspartate aminotransferase on substrate binding induces strain in the catalytic group and enhances catalysis.
|
| |
J Biol Chem,
278,
9481-9488.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Allert,
and
L.Baltzer
(2003).
Noncovalent binding of a reaction intermediate by a designed helix-loop-helix motif-implications for catalyst design.
|
| |
Chembiochem,
4,
306-318.
|
 |
|
|
|
|
 |
R.Omi,
M.Goto,
I.Miyahara,
H.Mizuguchi,
H.Hayashi,
H.Kagamiyama,
and
K.Hirotsu
(2003).
Crystal structures of threonine synthase from Thermus thermophilus HB8: conformational change, substrate recognition, and mechanism.
|
| |
J Biol Chem,
278,
46035-46045.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.Deu,
K.A.Koch,
and
J.F.Kirsch
(2002).
The role of the conserved Lys68*:Glu265 intersubunit salt bridge in aspartate aminotransferase kinetics: multiple forced covariant amino acid substitutions in natural variants.
|
| |
Protein Sci,
11,
1062-1073.
|
 |
|
|
|
|
 |
G.G.Hammes
(2002).
Multiple conformational changes in enzyme catalysis.
|
| |
Biochemistry,
41,
8221-8228.
|
 |
|
|
|
|
 |
K.Haruyama,
T.Nakai,
I.Miyahara,
K.Hirotsu,
H.Mizuguchi,
H.Hayashi,
and
H.Kagamiyama
(2001).
Structures of Escherichia coli histidinol-phosphate aminotransferase and its complexes with histidinol-phosphate and N-(5'-phosphopyridoxyl)-L-glutamate: double substrate recognition of the enzyme.
|
| |
Biochemistry,
40,
4633-4644.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
N.Yennawar,
J.Dunbar,
M.Conway,
S.Hutson,
and
G.Farber
(2001).
The structure of human mitochondrial branched-chain aminotransferase.
|
| |
Acta Crystallogr D Biol Crystallogr,
57,
506-515.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.M.Gulick,
B.K.Hubbard,
J.A.Gerlt,
and
I.Rayment
(2000).
Evolution of enzymatic activities in the enolase superfamily: crystallographic and mutagenesis studies of the reaction catalyzed by D-glucarate dehydratase from Escherichia coli.
|
| |
Biochemistry,
39,
4590-4602.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.D.Kern,
M.A.Oliveira,
P.Coffino,
and
M.L.Hackert
(1999).
Structure of mammalian ornithine decarboxylase at 1.6 A resolution: stereochemical implications of PLP-dependent amino acid decarboxylases.
|
| |
Structure,
7,
567-581.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
P.Storici,
G.Capitani,
D.De Biase,
M.Moser,
R.A.John,
J.N.Jansonius,
and
T.Schirmer
(1999).
Crystal structure of GABA-aminotransferase, a target for antiepileptic drug therapy.
|
| |
Biochemistry,
38,
8628-8634.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Azzariti,
R.A.Vacca,
S.Giannattasio,
R.S.Merafina,
E.Marra,
and
S.Doonan
(1998).
Kinetic properties and thermal stabilities of mutant forms of mitochondrial aspartate aminotransferase.
|
| |
Biochim Biophys Acta,
1386,
29-38.
|
 |
|
|
|
|
 |
D.Peisach,
D.M.Chipman,
P.W.Van Ophem,
J.M.Manning,
and
D.Ringe
(1998).
Crystallographic study of steps along the reaction pathway of D-amino acid aminotransferase.
|
| |
Biochemistry,
37,
4958-4967.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.Hayashi,
and
H.Kagamiyama
(1997).
Transient-state kinetics of the reaction of aspartate aminotransferase with aspartate at low pH reveals dual routes in the enzyme-substrate association process.
|
| |
Biochemistry,
36,
13558-13569.
|
 |
|
|
|
|
 |
R.A.Vacca,
S.Giannattasio,
R.Graber,
E.Sandmeier,
E.Marra,
and
P.Christen
(1997).
Active-site Arg --> Lys substitutions alter reaction and substrate specificity of aspartate aminotransferase.
|
| |
J Biol Chem,
272,
21932-21937.
|
 |
|
|
|
|
 |
Y.Park,
J.Luo,
P.G.Schultz,
and
J.F.Kirsch
(1997).
Noncoded amino acid replacement probes of the aspartate aminotransferase mechanism.
|
| |
Biochemistry,
36,
10517-10525.
|
 |
|
|
|
|
 |
A.G.von Stosch
(1996).
Aspartate aminotransferase complexed with erythro-beta-hydroxyaspartate: crystallographic and spectroscopic identification of the carbinolamine intermediate.
|
| |
Biochemistry,
35,
15260-15268.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.J.Adcock,
P.J.Gaskin,
P.N.Shaw,
P.H.Teesdale-Spittle,
and
L.D.Buckberry
(1996).
Novel sources of mammalian C-S lyase activity.
|
| |
J Pharm Pharmacol,
48,
150-153.
|
 |
|
|
|
|
 |
J.M.Goldberg,
and
J.F.Kirsch
(1996).
The reaction catalyzed by Escherichia coli aspartate aminotransferase has multiple partially rate-determining steps, while that catalyzed by the Y225F mutant is dominated by ketimine hydrolysis.
|
| |
Biochemistry,
35,
5280-5291.
|
 |
|
|
|
|
 |
M.Moser,
R.Müller,
N.Battchikova,
M.Koivulehto,
T.Korpela,
and
J.N.Jansonius
(1996).
Crystallization and preliminary X-ray analysis of phosphoserine aminotransferase from Bacillus circulans subsp. alkalophilus.
|
| |
Protein Sci,
5,
1426-1428.
|
 |
|
|
|
|
 |
Z.Marković-Housley,
T.Schirmer,
E.Hohenester,
A.R.Khomutov,
R.M.Khomutov,
M.Y.Karpeisky,
E.Sandmeier,
P.Christen,
and
J.N.Jansonius
(1996).
Crystal structures and solution studies of oxime adducts of mitochondrial aspartate aminotransferase.
|
| |
Eur J Biochem,
236,
1025-1032.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.J.Onuffer,
B.T.Ton,
I.Klement,
and
J.F.Kirsch
(1995).
The use of natural and unnatural amino acid substrates to define the substrate specificity differences of Escherichia coli aspartate and tyrosine aminotransferases.
|
| |
Protein Sci,
4,
1743-1749.
|
 |
|
|
|
|
 |
L.Birolo,
E.Sandmeier,
P.Christen,
and
R.A.John
(1995).
The roles of Tyr70 and Tyr225 in aspartate aminotransferase assessed by analysing the effects of mutations on the multiple reactions of the substrate analogue serine o-sulphate.
|
| |
Eur J Biochem,
232,
859-864.
|
 |
|
|
|
|
 |
V.N.Malashkevich,
J.J.Onuffer,
J.F.Kirsch,
and
J.N.Jansonius
(1995).
Alternating arginine-modulated substrate specificity in an engineered tyrosine aminotransferase.
|
| |
Nat Struct Biol,
2,
548-553.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.M.Duke,
S.Wakatsuki,
A.Hadfield,
and
L.N.Johnson
(1994).
Laue and monochromatic diffraction studies on catalysis in phosphorylase b crystals.
|
| |
Protein Sci,
3,
1178-1196.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

