 |
PDBsum entry 7odc

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.1.1.17
- ornithine decarboxylase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Spermine Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-ornithine + H+ = putrescine + CO2
|
 |
 |
 |
 |
 |
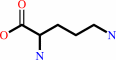 L-ornithine
L-ornithine
|
+
|
H(+)
|
=
|
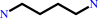 putrescine
putrescine
|
+
|
 CO2
CO2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
PLP)
matches with 93.75% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Structure
7:567-581
(1999)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure of mammalian ornithine decarboxylase at 1.6 A resolution: stereochemical implications of PLP-dependent amino acid decarboxylases.
|
|
A.D.Kern,
M.A.Oliveira,
P.Coffino,
M.L.Hackert.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
BACKGROUND: Pyridoxal-5'-phosphate (PLP) dependent enzymes catalyze a broad
range of reactions, resulting in bond cleavage at C alpha, C beta, or C gamma
carbons of D and L amino acid substrates. Ornithine decarboxylase (ODC) is a
PLP-dependent enzyme that controls a critical step in the biosynthesis of
polyamines, small organic polycations whose controlled levels are essential for
proper growth. ODC inhibition has applications for the treatment of certain
cancers and parasitic ailments such as African sleeping sickness. RESULTS: The
structure of truncated mouse ODC (mODC') was determined by multiple isomorphous
replacement methods and refined to 1.6 A resolution. This is the first structure
of a Group IV decarboxylase. The monomer contains two domains: an alpha/beta
barrel that binds the cofactor, and a second domain consisting mostly of beta
structure. Only the dimer is catalytically active, as the active sites are
constructed of residues from both monomers. The interactions stabilizing the
dimer shed light on its regulation by antizyme. The overall structure and the
environment of the cofactor are compared with those of alanine racemase.
CONCLUSIONS: The analysis of the mODC' structure and its comparison with alanine
racemase, together with modeling studies of the external aldimine intermediate,
provide insight into the stereochemical characteristics of PLP-dependent
decarboxylation. The structure comparison reveals stereochemical differences
with other PLP-dependent enzymes and the bacterial ODC. These characteristics
may be exploited in the design of new inhibitors specific for eukaryotic and
bacterial ODCs, and provide the basis for a detailed understanding of the
mechanism by which these enzymes regulate reaction specificity.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |
Figure 5.
Figure 5. Active site of mODC′ and comparison with ALR. (a)
Schematic drawing of the mODC′ active site illustrating the
hydrogen-bond interactions. Residues shown in bold face are
nearer the viewer. (b) Stereo figure of the active site of
mODC′ with electron density superimposed with its model. K69
of mODC′ is in Schiff-base linkage to the cofactor, E274 pairs
with the pyridine ring nitrogen N1, and H197 stacks on the si
face of the cofactor ring. Note the angle between K69 and the
pyridine ring of the cofactor exposing the si face. The map is a
2F[o]–F[c] map at 1.6 Å resolution contoured at 1.2σ.
(c) A view of the ALR and mODC′ active sites resulting from
the superposition of their cofactor rings. The mODC′ active
site is depicted in light gray. The figures were generated using
BOBSCRIPT [83], MOLSCRIPT [80] and Raster3D [81].
|
 |
|
|

|
| |
The above figure is
reprinted
by permission from Cell Press:
Structure
(1999,
7,
567-581)
copyright 1999.
|
|
| |
Figure was
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
G.Colotti,
and
A.Ilari
(2011).
Polyamine metabolism in Leishmania: from arginine to trypanothione.
|
| |
Amino Acids,
40,
269-285.
|
 |
|
|
|
|
 |
I.P.Ivanov,
A.E.Firth,
and
J.F.Atkins
(2010).
Recurrent emergence of catalytically inactive ornithine decarboxylase homologous forms that likely have regulatory function.
|
| |
J Mol Evol,
70,
289-302.
|
 |
|
|
|
|
 |
G.Steinkellner,
R.Rader,
G.G.Thallinger,
C.Kratky,
and
K.Gruber
(2009).
VASCo: computation and visualization of annotated protein surface contacts.
|
| |
BMC Bioinformatics,
10,
32.
|
 |
|
|
|
|
 |
K.L.Su,
Y.F.Liao,
H.C.Hung,
and
G.Y.Liu
(2009).
Critical factors determining dimerization of human antizyme inhibitor.
|
| |
J Biol Chem,
284,
26768-26777.
|
 |
|
|
|
|
 |
S.Weyand,
G.Kefala,
D.I.Svergun,
and
M.S.Weiss
(2009).
The three-dimensional structure of diaminopimelate decarboxylase from Mycobacterium tuberculosis reveals a tetrameric enzyme organisation.
|
| |
J Struct Funct Genomics,
10,
209-217.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.A.Moya-García,
J.Ruiz-Pernía,
S.Martí,
F.Sánchez-Jiménez,
and
I.Tuñón
(2008).
Analysis of the decarboxylation step in mammalian histidine decarboxylase. A computational study.
|
| |
J Biol Chem,
283,
12393-12401.
|
 |
|
|
|
|
 |
A.Jhingran,
P.K.Padmanabhan,
S.Singh,
K.Anamika,
A.A.Bakre,
S.Bhattacharya,
A.Bhattacharya,
N.Srinivasan,
and
R.Madhubala
(2008).
Characterization of the Entamoeba histolytica Ornithine Decarboxylase-Like Enzyme.
|
| |
PLoS Negl Trop Dis,
2,
e115.
|
 |
|
|
|
|
 |
I.Jariel-Encontre,
G.Bossis,
and
M.Piechaczyk
(2008).
Ubiquitin-independent degradation of proteins by the proteasome.
|
| |
Biochim Biophys Acta,
1786,
153-177.
|
 |
|
|
|
|
 |
S.Albeck,
O.Dym,
T.Unger,
Z.Snapir,
Z.Bercovich,
and
C.Kahana
(2008).
Crystallographic and biochemical studies revealing the structural basis for antizyme inhibitor function.
|
| |
Protein Sci,
17,
793-802.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.Hu,
D.Wu,
J.Chen,
J.Ding,
H.Jiang,
and
X.Shen
(2008).
The catalytic intermediate stabilized by a "down" active site loop for diaminopimelate decarboxylase from Helicobacter pylori. Enzymatic characterization with crystal structure analysis.
|
| |
J Biol Chem,
283,
21284-21293.
|
 |
|
|
|
|
 |
J.Lee,
A.J.Michael,
D.Martynowski,
E.J.Goldsmith,
and
M.A.Phillips
(2007).
Phylogenetic diversity and the structural basis of substrate specificity in the beta/alpha-barrel fold basic amino acid decarboxylases.
|
| |
J Biol Chem,
282,
27115-27125.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.Shah,
R.Akella,
E.J.Goldsmith,
and
M.A.Phillips
(2007).
X-ray structure of Paramecium bursaria Chlorella virus arginine decarboxylase: insight into the structural basis for substrate specificity.
|
| |
Biochemistry,
46,
2831-2841.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.A.Hoyt,
J.Zich,
J.Takeuchi,
M.Zhang,
C.Govaerts,
and
P.Coffino
(2006).
Glycine-alanine repeats impair proper substrate unfolding by the proteasome.
|
| |
EMBO J,
25,
1720-1729.
|
 |
|
|
|
|
 |
Y.Yamaguchi,
Y.Takatsuka,
S.Matsufuji,
Y.Murakami,
and
Y.Kamio
(2006).
Characterization of a counterpart to Mammalian ornithine decarboxylase antizyme in prokaryotes.
|
| |
J Biol Chem,
281,
3995-4001.
|
 |
|
|
|
|
 |
D.I.Dutyshev,
E.L.Darii,
N.P.Fomenkova,
I.V.Pechik,
K.M.Polyakov,
S.V.Nikonov,
N.S.Andreeva,
and
B.S.Sukhareva
(2005).
Structure of Escherichia coli glutamate decarboxylase (GADalpha) in complex with glutarate at 2.05 angstroms resolution.
|
| |
Acta Crystallogr D Biol Crystallogr,
61,
230-235.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.C.Eliot,
and
J.F.Kirsch
(2004).
Pyridoxal phosphate enzymes: mechanistic, structural, and evolutionary considerations.
|
| |
Annu Rev Biochem,
73,
383-415.
|
 |
|
|
|
|
 |
C.N.Patel,
R.S.Adcock,
K.G.Sell,
and
M.A.Oliveira
(2004).
Crystallization, X-ray diffraction and oligomeric characterization of arginine decarboxylase from Yersinia pestis, a key polyamine biosynthetic enzyme.
|
| |
Acta Crystallogr D Biol Crystallogr,
60,
2396-2398.
|
 |
|
|
|
|
 |
J.H.Lee,
M.Y.Son,
M.Y.Yoon,
J.D.Choi,
and
Y.T.Kim
(2004).
Isolation and characterization of ornithine decarboxylase gene from flounder (Paralichthys olivaceus).
|
| |
Mar Biotechnol (NY),
6,
453-462.
|
 |
|
|
|
|
 |
M.Zhang,
and
P.Coffino
(2004).
Repeat sequence of Epstein-Barr virus-encoded nuclear antigen 1 protein interrupts proteasome substrate processing.
|
| |
J Biol Chem,
279,
8635-8641.
|
 |
|
|
|
|
 |
R.Shah,
C.S.Coleman,
K.Mir,
J.Baldwin,
J.L.Van Etten,
N.V.Grishin,
A.E.Pegg,
B.A.Stanley,
and
M.A.Phillips
(2004).
Paramecium bursaria chlorella virus-1 encodes an unusual arginine decarboxylase that is a close homolog of eukaryotic ornithine decarboxylases.
|
| |
J Biol Chem,
279,
35760-35767.
|
 |
|
|
|
|
 |
Y.Takatsuka,
and
Y.Kamio
(2004).
Molecular dissection of the Selenomonas ruminantium cell envelope and lysine decarboxylase involved in the biosynthesis of a polyamine covalently linked to the cell wall peptidoglycan layer.
|
| |
Biosci Biotechnol Biochem,
68,
1.
|
 |
|
|
|
|
 |
B.E.Shakhnovich,
J.M.Harvey,
S.Comeau,
D.Lorenz,
C.DeLisi,
and
E.Shakhnovich
(2003).
ELISA: structure-function inferences based on statistically significant and evolutionarily inspired observations.
|
| |
BMC Bioinformatics,
4,
34.
|
 |
|
|
|
|
 |
C.V.Smith,
and
J.C.Sacchettini
(2003).
Mycobacterium tuberculosis: a model system for structural genomics.
|
| |
Curr Opin Struct Biol,
13,
658-664.
|
 |
|
|
|
|
 |
K.Gokulan,
B.Rupp,
M.S.Pavelka,
W.R.Jacobs,
and
J.C.Sacchettini
(2003).
Crystal structure of Mycobacterium tuberculosis diaminopimelate decarboxylase, an essential enzyme in bacterial lysine biosynthesis.
|
| |
J Biol Chem,
278,
18588-18596.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.Birkholtz,
F.Joubert,
A.W.Neitz,
and
A.I.Louw
(2003).
Comparative properties of a three-dimensional model of Plasmodium falciparum ornithine decarboxylase.
|
| |
Proteins,
50,
464-473.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
L.K.Jackson,
E.J.Goldsmith,
and
M.A.Phillips
(2003).
X-ray structure determination of Trypanosoma brucei ornithine decarboxylase bound to D-ornithine and to G418: insights into substrate binding and ODC conformational flexibility.
|
| |
J Biol Chem,
278,
22037-22043.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Zhang,
C.M.Pickart,
and
P.Coffino
(2003).
Determinants of proteasome recognition of ornithine decarboxylase, a ubiquitin-independent substrate.
|
| |
EMBO J,
22,
1488-1496.
|
 |
|
|
|
|
 |
P.B.Balbo,
C.N.Patel,
K.G.Sell,
R.S.Adcock,
S.Neelakantan,
P.A.Crooks,
and
M.A.Oliveira
(2003).
Spectrophotometric and steady-state kinetic analysis of the biosynthetic arginine decarboxylase of Yersinia pestis utilizing arginine analogues as inhibitors and alternative substrates.
|
| |
Biochemistry,
42,
15189-15196.
|
 |
|
|
|
|
 |
S.Eswaramoorthy,
S.Gerchman,
V.Graziano,
H.Kycia,
F.W.Studier,
and
S.Swaminathan
(2003).
Structure of a yeast hypothetical protein selected by a structural genomics approach.
|
| |
Acta Crystallogr D Biol Crystallogr,
59,
127-135.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.Momany,
V.Levdikov,
L.Blagova,
and
K.Crews
(2002).
Crystallization of diaminopimelate decarboxylase from Escherichia coli, a stereospecific D-amino-acid decarboxylase.
|
| |
Acta Crystallogr D Biol Crystallogr,
58,
549-552.
|
 |
|
|
|
|
 |
H.Chen,
A.MacDonald,
and
P.Coffino
(2002).
Structural elements of antizymes 1 and 2 are required for proteasomal degradation of ornithine decarboxylase.
|
| |
J Biol Chem,
277,
45957-45961.
|
 |
|
|
|
|
 |
C.Hanfrey,
S.Sommer,
M.J.Mayer,
D.Burtin,
and
A.J.Michael
(2001).
Arabidopsis polyamine biosynthesis: absence of ornithine decarboxylase and the mechanism of arginine decarboxylase activity.
|
| |
Plant J,
27,
551-560.
|
 |
|
|
|
|
 |
H.Kagamiyama,
and
H.Hayashi
(2001).
Release of enzyme strain during catalysis reduces the activation energy barrier.
|
| |
Chem Rec,
1,
385-394.
|
 |
|
|
|
|
 |
P.Christen,
and
P.K.Mehta
(2001).
From cofactor to enzymes. The molecular evolution of pyridoxal-5'-phosphate-dependent enzymes.
|
| |
Chem Rec,
1,
436-447.
|
 |
|
|
|
|
 |
G.Schneider,
H.Käck,
and
Y.Lindqvist
(2000).
The manifold of vitamin B6 dependent enzymes.
|
| |
Structure,
8,
R1-R6.
|
 |
|
|
|
|
 |
P.Coffino
(2000).
Polyamines in spermiogenesis: not now, darling.
|
| |
Proc Natl Acad Sci U S A,
97,
4421-4423.
|
 |
|
|
|
|
 |
Y.Takatsuka,
Y.Yamaguchi,
M.Ono,
and
Y.Kamio
(2000).
Gene cloning and molecular characterization of lysine decarboxylase from Selenomonas ruminantium delineate its evolutionary relationship to ornithine decarboxylases from eukaryotes.
|
| |
J Bacteriol,
182,
6732-6741.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

