
|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
Chains A, B:
E.C.2.5.1.58
- protein farnesyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-cysteinyl-[protein] + (2E,6E)-farnesyl diphosphate = S-(2E,6E)- farnesyl-L-cysteinyl-[protein] + diphosphate
|
 |
 |
 |
 |
 |
L-cysteinyl-[protein]
|
+
|
 (2E,6E)-farnesyl diphosphate
(2E,6E)-farnesyl diphosphate
|
=
|
S-(2E,6E)- farnesyl-L-cysteinyl-[protein]
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Mg(2+); Zn(2+)
|
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
Chain A:
E.C.2.5.1.59
- protein geranylgeranyltransferase type I.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
geranylgeranyl diphosphate + L-cysteinyl-[protein] = S-geranylgeranyl-L- cysteinyl-[protein] + diphosphate
|
 |
 |
 |
 |
 |
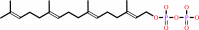 geranylgeranyl diphosphate
geranylgeranyl diphosphate
|
+
|
L-cysteinyl-[protein]
|
=
|
S-geranylgeranyl-L- cysteinyl-[protein]
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Zn(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Science
275:1800-1804
(1997)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of protein farnesyltransferase at 2.25 angstrom resolution.
|
|
H.W.Park,
S.R.Boduluri,
J.F.Moomaw,
P.J.Casey,
L.S.Beese.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Protein farnesyltransferase (FTase) catalyzes the carboxyl-terminal lipidation
of Ras and several other cellular signal transduction proteins. The essential
nature of this modification for proper function of these proteins has led to the
emergence of FTase as a target for the development of new anticancer therapy.
Inhibition of this enzyme suppresses the transformed phenotype in cultured cells
and causes tumor regression in animal models. The crystal structure of
heterodimeric mammalian FTase was determined at 2.25 angstrom resolution. The
structure shows a combination of two unusual domains: a crescent-shaped
seven-helical hairpin domain and an alpha-alpha barrel domain. The active site
is formed by two clefts that intersect at a bound zinc ion. One cleft contains a
nine-residue peptide that may mimic the binding of the Ras substrate; the other
cleft is lined with highly conserved aromatic residues appropriate for binding
the farnesyl isoprenoid with required specificity.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Fig. 2. The  subunit.
(A) Aromatic pocket in the center of the subunit.
(A) Aromatic pocket in the center of the  - -  barrel of
the barrel of
the  subunit.
This view is a 90° clockwise^ rotation relative to Fig. 1A.
Only helices 2 subunit.
This view is a 90° clockwise^ rotation relative to Fig. 1A.
Only helices 2  to 13 to 13  are shown.
Yellow, the nine aromatic residues that line the pocket;
magenta, the zinc ion [MOLSCRIPT (40) and RASTER3D (41)]. (B) A
portion of the solvent-accessible surface showing some of the^
aromatic residues that line the putative FPP binding pocket. FPP
is modeled with the isoprenoid in the hydrophobic cleft and the^
diphosphate moiety positioned near the zinc. The carbon atoms of
FPP are yellow, oxygens are red, and phosphates are green. The
program INSIGHT II (43) was used to construct an
energy-minimized^ model of FPP and GRASP (44) was used to
calculate the accessible^ surface. are shown.
Yellow, the nine aromatic residues that line the pocket;
magenta, the zinc ion [MOLSCRIPT (40) and RASTER3D (41)]. (B) A
portion of the solvent-accessible surface showing some of the^
aromatic residues that line the putative FPP binding pocket. FPP
is modeled with the isoprenoid in the hydrophobic cleft and the^
diphosphate moiety positioned near the zinc. The carbon atoms of
FPP are yellow, oxygens are red, and phosphates are green. The
program INSIGHT II (43) was used to construct an
energy-minimized^ model of FPP and GRASP (44) was used to
calculate the accessible^ surface.
|
 |
Figure 3.
Fig. 3. (A) Solvent-accessible surface and electrostatic
surface potential. The dashed box highlights the cleft where
the^ nonapeptide binds. The most negative electrostatic surface
potential (-10 kT) is colored red. The most positive
electrostatic surface^ potential (10 kT) is blue. The
orientation is similar to that of Fig. 1. The arrow indicates
the putative FPP binding site [GRASP (44)]. (B) Close-up view of
the nonapeptide binding cleft bounded by the dashed lines in
(A). The COOH-terminus and six residues of the nonapeptide
(Ala^9-Val8-Thr7-Ser6-Asp5-Pro4) are visible. Atom colors for
the nonapeptide are coral, carbons; red, oxygen; light blue,
nitrogen; and zinc, magenta. (C) Stereo view of the nonapeptide
(COOH-terminus of a symmetry-related^  subunit).
The nonapeptide is numbered from the COOH-terminus. Atom colors
in the nonapeptide are coral, carbons; orange, oxygen; and light
blue, nitrogen. Atom colors of residues forming the^ binding
pocket are khaki, carbons; red, oxygen; and blue, nitrogen. Zinc
is a magenta sphere. Water molecules are red spheres. Dotted^
lines represent potential hydrogen bonds. subunit).
The nonapeptide is numbered from the COOH-terminus. Atom colors
in the nonapeptide are coral, carbons; orange, oxygen; and light
blue, nitrogen. Atom colors of residues forming the^ binding
pocket are khaki, carbons; red, oxygen; and blue, nitrogen. Zinc
is a magenta sphere. Water molecules are red spheres. Dotted^
lines represent potential hydrogen bonds.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the AAAs:
Science
(1997,
275,
1800-1804)
copyright 1997.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
N.Berndt,
A.D.Hamilton,
and
S.M.Sebti
(2011).
Targeting protein prenylation for cancer therapy.
|
| |
Nat Rev Cancer,
11,
775-791.
|
 |
|
|
|
|
 |
Y.Qiao,
J.Gao,
Y.Qiu,
L.Wu,
F.Guo,
K.K.Lo,
and
D.Li
(2011).
Design, synthesis, and characterization of piperazinedione-based dual protein inhibitors for both farnesyltransferase and geranylgeranyltransferase-I.
|
| |
Eur J Med Chem,
46,
2264-2273.
|
 |
|
|
|
|
 |
M.Andrews,
D.H.Huizinga,
and
D.N.Crowell
(2010).
The CaaX specificities of Arabidopsis protein prenyltransferases explain era1 and ggb phenotypes.
|
| |
BMC Plant Biol,
10,
118.
|
 |
|
|
|
|
 |
U.T.Nguyen,
R.S.Goody,
and
K.Alexandrov
(2010).
Understanding and exploiting protein prenyltransferases.
|
| |
Chembiochem,
11,
1194-1201.
|
 |
|
|
|
|
 |
O.Henry,
F.Lopez-Gallego,
S.A.Agger,
C.Schmidt-Dannert,
S.Sen,
D.Shintani,
K.Cornish,
and
M.D.Distefano
(2009).
A versatile photoactivatable probe designed to label the diphosphate binding site of farnesyl diphosphate utilizing enzymes.
|
| |
Bioorg Med Chem,
17,
4797-4805.
|
 |
|
|
|
|
 |
S.F.Sousa,
P.A.Fernandes,
and
M.J.Ramos
(2009).
The search for the mechanism of the reaction catalyzed by farnesyltransferase.
|
| |
Chemistry,
15,
4243-4247.
|
 |
|
|
|
|
 |
W.Zhang,
L.Wang,
Y.Liu,
J.Xu,
G.Zhu,
H.Cang,
X.Li,
M.Bartlam,
K.Hensley,
G.Li,
Z.Rao,
and
X.C.Zhang
(2009).
Structure of human lanthionine synthetase C-like protein 1 and its interaction with Eps8 and glutathione.
|
| |
Genes Dev,
23,
1387-1392.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.W.Christianson
(2008).
Unearthing the roots of the terpenome.
|
| |
Curr Opin Chem Biol,
12,
141-150.
|
 |
|
|
|
|
 |
M.A.Hast,
and
L.S.Beese
(2008).
Structure of protein geranylgeranyltransferase-I from the human pathogen Candida albicans complexed with a lipid substrate.
|
| |
J Biol Chem,
283,
31933-31940.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Koutmos,
R.Pejchal,
T.M.Bomer,
R.G.Matthews,
J.L.Smith,
and
M.L.Ludwig
(2008).
Metal active site elasticity linked to activation of homocysteine in methionine synthases.
|
| |
Proc Natl Acad Sci U S A,
105,
3286-3291.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
T.Subramanian,
S.Liu,
J.M.Troutman,
D.A.Andres,
and
H.P.Spielmann
(2008).
Protein farnesyltransferase-catalyzed isoprenoid transfer to peptide depends on lipid size and shape, not hydrophobicity.
|
| |
Chembiochem,
9,
2872-2882.
|
 |
|
|
|
|
 |
U.A.Mirza,
G.Chen,
Y.H.Liu,
R.J.Doll,
V.M.Girijavallabhan,
A.K.Ganguly,
and
B.N.Pramanik
(2008).
Mass spectrometric studies of potent inhibitors of farnesyl protein transferase--detection of pentameric noncovalent complexes.
|
| |
J Mass Spectrom,
43,
1393-1401.
|
 |
|
|
|
|
 |
B.Tamames,
S.F.Sousa,
J.Tamames,
P.A.Fernandes,
and
M.J.Ramos
(2007).
Analysis of zinc-ligand bond lengths in metalloproteins: trends and patterns.
|
| |
Proteins,
69,
466-475.
|
 |
|
|
|
|
 |
C.Wang,
Q.Tian,
Z.Hou,
M.Mucha,
M.Aukerman,
and
O.A.Olsen
(2007).
The Arabidopsis thaliana AT PRP39-1 gene, encoding a tetratricopeptide repeat protein with similarity to the yeast pre-mRNA processing protein PRP39, affects flowering time.
|
| |
Plant Cell Rep,
26,
1357-1366.
|
 |
|
|
|
|
 |
G.Cui,
and
K.M.Merz
(2007).
Computational studies of the farnesyltransferase ternary complex part II: the conformational activation of farnesyldiphosphate.
|
| |
Biochemistry,
46,
12375-12381.
|
 |
|
|
|
|
 |
J.Penner-Hahn
(2007).
Zinc-promoted alkyl transfer: a new role for zinc.
|
| |
Curr Opin Chem Biol,
11,
166-171.
|
 |
|
|
|
|
 |
M.Paul,
G.C.Patton,
and
W.A.van der Donk
(2007).
Mutants of the zinc ligands of lacticin 481 synthetase retain dehydration activity but have impaired cyclization activity.
|
| |
Biochemistry,
46,
6268-6276.
|
 |
|
|
|
|
 |
R.Rasteiro,
and
J.B.Pereira-Leal
(2007).
Multiple domain insertions and losses in the evolution of the Rab prenylation complex.
|
| |
BMC Evol Biol,
7,
140.
|
 |
|
|
|
|
 |
T.Raz,
V.Nardi,
M.Azam,
J.Cortes,
and
G.Q.Daley
(2007).
Farnesyl transferase inhibitor resistance probed by target mutagenesis.
|
| |
Blood,
110,
2102-2109.
|
 |
|
|
|
|
 |
W.Xie,
C.Zhou,
and
R.H.Huang
(2007).
Structure of tRNA dimethylallyltransferase: RNA modification through a channel.
|
| |
J Mol Biol,
367,
872-881.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.Bai,
T.C.Auperin,
C.Y.Chou,
G.G.Chang,
J.L.Manley,
and
L.Tong
(2007).
Crystal structure of murine CstF-77: dimeric association and implications for polyadenylation of mRNA precursors.
|
| |
Mol Cell,
25,
863-875.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
G.Hao,
B.Derakhshan,
L.Shi,
F.Campagne,
and
S.S.Gross
(2006).
SNOSID, a proteomic method for identification of cysteine S-nitrosylation sites in complex protein mixtures.
|
| |
Proc Natl Acad Sci U S A,
103,
1012-1017.
|
 |
|
|
|
|
 |
Y.Nakase,
K.Fukuda,
Y.Chikashige,
C.Tsutsumi,
D.Morita,
S.Kawamoto,
M.Ohnuki,
Y.Hiraoka,
and
T.Matsumoto
(2006).
A defect in protein farnesylation suppresses a loss of Schizosaccharomyces pombe tsc2+, a homolog of the human gene predisposing to tuberous sclerosis complex.
|
| |
Genetics,
173,
569-578.
|
 |
|
|
|
|
 |
G.Cui,
B.Wang,
and
K.M.Merz
(2005).
Computational studies of the farnesyltransferase ternary complex part I: substrate binding.
|
| |
Biochemistry,
44,
16513-16523.
|
 |
|
|
|
|
 |
N.Ferri,
R.Paoletti,
and
A.Corsini
(2005).
Lipid-modified proteins as biomarkers for cardiovascular disease: a review.
|
| |
Biomarkers,
10,
219-237.
|
 |
|
|
|
|
 |
S.F.Sousa,
P.A.Fernandes,
and
M.J.Ramos
(2005).
Farnesyltransferase--new insights into the zinc-coordination sphere paradigm: evidence for a carboxylate-shift mechanism.
|
| |
Biophys J,
88,
483-494.
|
 |
|
|
|
|
 |
T.Kuzuyama,
J.P.Noel,
and
S.B.Richard
(2005).
Structural basis for the promiscuous biosynthetic prenylation of aromatic natural products.
|
| |
Nature,
435,
983-987.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
W.C.Guida,
A.D.Hamilton,
J.W.Crotty,
and
S.M.Sebti
(2005).
Protein farnesyltransferase: flexible docking studies on inhibitors using computational modeling.
|
| |
J Comput Aided Mol Des,
19,
871-885.
|
 |
|
|
|
|
 |
W.Kou,
H.S.Kolla,
A.Ortiz-Acevedo,
D.C.Haines,
M.Junker,
and
G.R.Dieckmann
(2005).
Modulation of zinc- and cobalt-binding affinities through changes in the stability of the zinc ribbon protein L36.
|
| |
J Biol Inorg Chem,
10,
167-180.
|
 |
|
|
|
|
 |
K.Bracha-Drori,
K.Shichrur,
A.Katz,
M.Oliva,
R.Angelovici,
S.Yalovsky,
and
N.Ohad
(2004).
Detection of protein-protein interactions in plants using bimolecular fluorescence complementation.
|
| |
Plant J,
40,
419-427.
|
 |
|
|
|
|
 |
S.Y.Chang,
T.P.Ko,
A.P.Chen,
A.H.Wang,
and
P.H.Liang
(2004).
Substrate binding mode and reaction mechanism of undecaprenyl pyrophosphate synthase deduced from crystallographic studies.
|
| |
Protein Sci,
13,
971-978.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.S.Taylor,
T.S.Reid,
K.L.Terry,
P.J.Casey,
and
L.S.Beese
(2003).
Structure of mammalian protein geranylgeranyltransferase type-I.
|
| |
EMBO J,
22,
5963-5974.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Maurer-Stroh,
S.Washietl,
and
F.Eisenhaber
(2003).
Protein prenyltransferases.
|
| |
Genome Biol,
4,
212.
|
 |
|
|
|
|
 |
D.A.Whittington,
M.L.Wise,
M.Urbansky,
R.M.Coates,
R.B.Croteau,
and
D.W.Christianson
(2002).
Bornyl diphosphate synthase: structure and strategy for carbocation manipulation by a terpenoid cyclase.
|
| |
Proc Natl Acad Sci U S A,
99,
15375-15380.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.C.Evans,
D.P.Huddler,
J.Jiracek,
C.Castro,
N.S.Millian,
T.A.Garrow,
and
M.L.Ludwig
(2002).
Betaine-homocysteine methyltransferase: zinc in a distorted barrel.
|
| |
Structure,
10,
1159-1171.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
K.N.Cho,
and
K.I.Lee
(2002).
Chemistry and biology of Ras farnesyltransferase.
|
| |
Arch Pharm Res,
25,
759-769.
|
 |
|
|
|
|
 |
M.A.Huntley,
and
G.B.Golding
(2002).
Simple sequences are rare in the Protein Data Bank.
|
| |
Proteins,
48,
134-140.
|
 |
|
|
|
|
 |
P.H.Liang,
T.P.Ko,
and
A.H.Wang
(2002).
Structure, mechanism and function of prenyltransferases.
|
| |
Eur J Biochem,
269,
3339-3354.
|
 |
|
|
|
|
 |
S.B.Long,
P.J.Casey,
and
L.S.Beese
(2002).
Reaction path of protein farnesyltransferase at atomic resolution.
|
| |
Nature,
419,
645-650.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
X.X.Dong,
M.Ospeck,
and
K.H.Iwasa
(2002).
Piezoelectric reciprocal relationship of the membrane motor in the cochlear outer hair cell.
|
| |
Biophys J,
82,
1254-1259.
|
 |
|
|
|
|
 |
C.Steegborn,
O.Danot,
R.Huber,
and
T.Clausen
(2001).
Crystal structure of transcription factor MalT domain III: a novel helix repeat fold implicated in regulated oligomerization.
|
| |
Structure,
9,
1051-1060.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
H.C.Schmid,
U.Oster,
J.Kögel,
S.Lenz,
and
W.Rüdiger
(2001).
Cloning and characterisation of chlorophyll synthase from Avena sativa.
|
| |
Biol Chem,
382,
903-911.
|
 |
|
|
|
|
 |
K.E.Hightower,
P.J.Casey,
and
C.A.Fierke
(2001).
Farnesylation of nonpeptidic thiol compounds by protein farnesyltransferase.
|
| |
Biochemistry,
40,
1002-1010.
|
 |
|
|
|
|
 |
K.H.Iwasa
(2001).
A two-state piezoelectric model for outer hair cell motility.
|
| |
Biophys J,
81,
2495-2506.
|
 |
|
|
|
|
 |
M.Crul,
G.J.de Klerk,
J.H.Beijnen,
and
J.H.Schellens
(2001).
Ras biochemistry and farnesyl transferase inhibitors: a literature survey.
|
| |
Anticancer Drugs,
12,
163-184.
|
 |
|
|
|
|
 |
M.Fujihashi,
Y.W.Zhang,
Y.Higuchi,
X.Y.Li,
T.Koyama,
and
K.Miki
(2001).
Crystal structure of cis-prenyl chain elongating enzyme, undecaprenyl diphosphate synthase.
|
| |
Proc Natl Acad Sci U S A,
98,
4337-4342.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.Roskoski,
and
P.A.Ritchie
(2001).
Time-dependent inhibition of protein farnesyltransferase by a benzodiazepine peptide mimetic.
|
| |
Biochemistry,
40,
9329-9335.
|
 |
|
|
|
|
 |
S.B.Long,
P.J.Hancock,
A.M.Kral,
H.W.Hellinga,
and
L.S.Beese
(2001).
The crystal structure of human protein farnesyltransferase reveals the basis for inhibition by CaaX tetrapeptides and their mimetics.
|
| |
Proc Natl Acad Sci U S A,
98,
12948-12953.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
X.Wang,
P.D.Zamore,
and
T.M.Hall
(2001).
Crystal structure of a Pumilio homology domain.
|
| |
Mol Cell,
7,
855-865.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
B.Kobe,
and
A.V.Kajava
(2000).
When protein folding is simplified to protein coiling: the continuum of solenoid protein structures.
|
| |
Trends Biochem Sci,
25,
509-515.
|
 |
|
|
|
|
 |
C.Huang,
K.E.Hightower,
and
C.A.Fierke
(2000).
Mechanistic studies of rat protein farnesyltransferase indicate an associative transition state.
|
| |
Biochemistry,
39,
2593-2602.
|
 |
|
|
|
|
 |
C.Z.Ding,
J.T.Hunt,
C.Ricca,
and
V.Manne
(2000).
3-Imidazolylmethylaminophenylsulfonyltetrahydroquinolines, a novel series of farnesyltransferase inhibitors.
|
| |
Bioorg Med Chem Lett,
10,
273-275.
|
 |
|
|
|
|
 |
E.C.Ziegelhoffer,
L.J.Medrano,
and
E.M.Meyerowitz
(2000).
Cloning of the Arabidopsis WIGGUM gene identifies a role for farnesylation in meristem development.
|
| |
Proc Natl Acad Sci U S A,
97,
7633-7638.
|
 |
|
|
|
|
 |
F.S.Buckner,
K.Yokoyama,
L.Nguyen,
A.Grewal,
H.Erdjument-Bromage,
P.Tempst,
C.L.Strickland,
L.Xiao,
W.C.Van Voorhis,
and
M.H.Gelb
(2000).
Cloning, heterologous expression, and distinct substrate specificity of protein farnesyltransferase from Trypanosoma brucei.
|
| |
J Biol Chem,
275,
21870-21876.
|
 |
|
|
|
|
 |
F.Vallée,
F.Lipari,
P.Yip,
B.Sleno,
A.Herscovics,
and
P.L.Howell
(2000).
Crystal structure of a class I alpha1,2-mannosidase involved in N-glycan processing and endoplasmic reticulum quality control.
|
| |
EMBO J,
19,
581-588.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
G.C.Prendergast,
and
A.Oliff
(2000).
Farnesyltransferase inhibitors: antineoplastic properties, mechanisms of action, and clinical prospects.
|
| |
Semin Cancer Biol,
10,
443-452.
|
 |
|
|
|
|
 |
H.Zhang,
M.C.Seabra,
and
J.Deisenhofer
(2000).
Crystal structure of Rab geranylgeranyltransferase at 2.0 A resolution.
|
| |
Structure,
8,
241-251.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.T.Stickney,
and
J.E.Buss
(2000).
Murine guanylate-binding protein: incomplete geranylgeranyl isoprenoid modification of an interferon-gamma-inducible guanosine triphosphate-binding protein.
|
| |
Mol Biol Cell,
11,
2191-2200.
|
 |
|
|
|
|
 |
K.Sauer,
and
R.K.Thauer
(2000).
Methyl-coenzyme M formation in methanogenic archaea. Involvement of zinc in coenzyme M activation.
|
| |
Eur J Biochem,
267,
2498-2504.
|
 |
|
|
|
|
 |
M.J.Saderholm,
K.E.Hightower,
and
C.A.Fierke
(2000).
Role of metals in the reaction catalyzed by protein farnesyltransferase.
|
| |
Biochemistry,
39,
12398-12405.
|
 |
|
|
|
|
 |
R.A.Spence,
K.E.Hightower,
K.L.Terry,
L.S.Beese,
C.A.Fierke,
and
P.J.Casey
(2000).
Conversion of Tyr361 beta to Leu in mammalian protein farnesyltransferase impairs product release but not substrate recognition.
|
| |
Biochemistry,
39,
13651-13659.
|
 |
|
|
|
|
 |
S.B.Long,
P.J.Casey,
and
L.S.Beese
(2000).
The basis for K-Ras4B binding specificity to protein farnesyltransferase revealed by 2 A resolution ternary complex structures.
|
| |
Structure,
8,
209-222.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.P.Pang,
K.Xu,
J.E.Yazal,
and
F.G.Prendergas
(2000).
Successful molecular dynamics simulation of the zinc-bound farnesyltransferase using the cationic dummy atom approach.
|
| |
Protein Sci,
9,
1857-1865.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Oliff
(1999).
Farnesyltransferase inhibitors: targeting the molecular basis of cancer.
|
| |
Biochim Biophys Acta,
1423,
C19-C30.
|
 |
|
|
|
|
 |
B.Kobe,
T.Gleichmann,
J.Horne,
I.G.Jennings,
P.D.Scotney,
and
T.Teh
(1999).
Turn up the HEAT.
|
| |
Structure,
7,
R91-R97.
|
 |
|
|
|
|
 |
H.Kim,
and
C.H.Yang
(1999).
Active site determination of yeast geranylgeranyl protein transferase type I expressed in Escherichia coli.
|
| |
Eur J Biochem,
265,
105-111.
|
 |
|
|
|
|
 |
H.Zhang,
and
N.V.Grishin
(1999).
The alpha-subunit of protein prenyltransferases is a member of the tetratricopeptide repeat family.
|
| |
Protein Sci,
8,
1658-1667.
|
 |
|
|
|
|
 |
J.A.Boutin,
W.Marande,
L.Petit,
A.Loynel,
C.Desmet,
E.Canet,
and
J.L.Fauchère
(1999).
Investigation of S-farnesyl transferase substrate specificity with combinatorial tetrapeptide libraries.
|
| |
Cell Signal,
11,
59-69.
|
 |
|
|
|
|
 |
K.E.Hightower,
and
C.A.Fierke
(1999).
Zinc-catalyzed sulfur alkyation:insights from protein farnesyltransferase.
|
| |
Curr Opin Chem Biol,
3,
176-181.
|
 |
|
|
|
|
 |
M.R.Groves,
and
D.Barford
(1999).
Topological characteristics of helical repeat proteins.
|
| |
Curr Opin Struct Biol,
9,
383-389.
|
 |
|
|
|
|
 |
M.R.Groves,
N.Hanlon,
P.Turowski,
B.A.Hemmings,
and
D.Barford
(1999).
The structure of the protein phosphatase 2A PR65/A subunit reveals the conformation of its 15 tandemly repeated HEAT motifs.
|
| |
Cell,
96,
99.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.M.Ciccarone,
S.C.MacTough,
T.M.Williams,
C.J.Dinsmore,
T.J.O'Neill,
D.Shah,
J.C.Culberson,
K.S.Koblan,
N.E.Kohl,
J.B.Gibbs,
A.I.Oliff,
S.L.Graham,
and
G.D.Hartman
(1999).
Non-thiol 3-aminomethylbenzamide inhibitors of farnesyl-protein transferase.
|
| |
Bioorg Med Chem Lett,
9,
1991-1996.
|
 |
|
|
|
|
 |
Z.S.Zhou,
K.Peariso,
J.E.Penner-Hahn,
and
R.G.Matthews
(1999).
Identification of the zinc ligands in cobalamin-independent methionine synthase (MetE) from Escherichia coli.
|
| |
Biochemistry,
38,
15915-15926.
|
 |
|
|
|
|
 |
C.L.Strickland,
W.T.Windsor,
R.Syto,
L.Wang,
R.Bond,
Z.Wu,
J.Schwartz,
H.V.Le,
L.S.Beese,
and
P.C.Weber
(1998).
Crystal structure of farnesyl protein transferase complexed with a CaaX peptide and farnesyl diphosphate analogue.
|
| |
Biochemistry,
37,
16601-16611.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.T.Pool,
and
T.E.Thompson
(1998).
Chain length and temperature dependence of the reversible association of model acylated proteins with lipid bilayers.
|
| |
Biochemistry,
37,
10246-10255.
|
 |
|
|
|
|
 |
D.C.Heimbrook,
and
A.Oliff
(1998).
Therapeutic intervention and signaling.
|
| |
Curr Opin Cell Biol,
10,
284-288.
|
 |
|
|
|
|
 |
H.W.Fu,
L.S.Beese,
and
P.J.Casey
(1998).
Kinetic analysis of zinc ligand mutants of mammalian protein farnesyltransferase.
|
| |
Biochemistry,
37,
4465-4472.
|
 |
|
|
|
|
 |
J.E.Coleman
(1998).
Zinc enzymes.
|
| |
Curr Opin Chem Biol,
2,
222-234.
|
 |
|
|
|
|
 |
J.S.Anant,
L.Desnoyers,
M.Machius,
B.Demeler,
J.C.Hansen,
K.D.Westover,
J.Deisenhofer,
and
M.C.Seabra
(1998).
Mechanism of Rab geranylgeranylation: formation of the catalytic ternary complex.
|
| |
Biochemistry,
37,
12559-12568.
|
 |
|
|
|
|
 |
K.E.Hightower,
C.C.Huang,
P.J.Casey,
and
C.A.Fierke
(1998).
H-Ras peptide and protein substrates bind protein farnesyltransferase as an ionized thiolate.
|
| |
Biochemistry,
37,
15555-15562.
|
 |
|
|
|
|
 |
K.U.Wendt,
and
G.E.Schulz
(1998).
Isoprenoid biosynthesis: manifold chemistry catalyzed by similar enzymes.
|
| |
Structure,
6,
127-133.
|
 |
|
|
|
|
 |
L.Desnoyers,
and
M.C.Seabra
(1998).
Single prenyl-binding site on protein prenyl transferases.
|
| |
Proc Natl Acad Sci U S A,
95,
12266-12270.
|
 |
|
|
|
|
 |
M.H.Gelb,
J.D.Scholten,
and
J.S.Sebolt-Leopold
(1998).
Protein prenylation: from discovery to prospects for cancer treatment.
|
| |
Curr Opin Chem Biol,
2,
40-48.
|
 |
|
|
|
|
 |
M.Schlitzer
(1998).
[Inhibitors of farnesyltransferase: a new approach for development of potential cancer drugs]
|
| |
Pharm Unserer Zeit,
27,
278-288.
|
 |
|
|
|
|
 |
P.Dunten,
U.Kammlott,
R.Crowther,
D.Weber,
R.Palermo,
and
J.Birktoft
(1998).
Protein farnesyltransferase: structure and implications for substrate binding.
|
| |
Biochemistry,
37,
7907-7912.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.G.Kibbey,
J.Rizo,
L.M.Gierasch,
and
R.G.Anderson
(1998).
The LDL receptor clustering motif interacts with the clathrin terminal domain in a reverse turn conformation.
|
| |
J Cell Biol,
142,
59-67.
|
 |
|
|
|
|
 |
S.B.Long,
P.J.Casey,
and
L.S.Beese
(1998).
Cocrystal structure of protein farnesyltransferase complexed with a farnesyl diphosphate substrate.
|
| |
Biochemistry,
37,
9612-9618.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.E.Trueblood,
V.L.Boyartchuk,
and
J.Rine
(1997).
Substrate specificity determinants in the farnesyltransferase beta-subunit.
|
| |
Proc Natl Acad Sci U S A,
94,
10774-10779.
|
 |
|
|
|
|
 |
H.W.Park,
and
L.S.Beese
(1997).
Protein farnesyltransferase.
|
| |
Curr Opin Struct Biol,
7,
873-880.
|
 |
|
|
|
|
 |
J.B.Gibbs,
S.L.Graham,
G.D.Hartman,
K.S.Koblan,
N.E.Kohl,
C.A.Omer,
and
A.Oliff
(1997).
Farnesyltransferase inhibitors versus Ras inhibitors.
|
| |
Curr Opin Chem Biol,
1,
197-203.
|
 |
|
|
|
|
 |
J.M.Dolence,
D.B.Rozema,
and
C.D.Poulter
(1997).
Yeast protein farnesyltransferase. Site-directed mutagenesis of conserved residues in the beta-subunit.
|
| |
Biochemistry,
36,
9246-9252.
|
 |
|
|
|
|
 |
M.Geyer,
and
A.Wittinghofer
(1997).
GEFs, GAPs, GDIs and effectors: taking a closer (3D) look at the regulation of Ras-related GTP-binding proteins.
|
| |
Curr Opin Struct Biol,
7,
786-792.
|
 |
|
|
|
|
 |
R.G.Matthews,
and
C.W.Goulding
(1997).
Enzyme-catalyzed methyl transfers to thiols: the role of zinc.
|
| |
Curr Opin Chem Biol,
1,
332-339.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

