 |
PDBsum entry 1v7u

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Crystal structure of undecaprenyl pyrophosphate synthase with farnesyl pyrophosphate
|
|
Structure:
|
 |
Undecaprenyl pyrophosphate synthetase. Chain: a, b. Synonym: upp synthetase, di-trans-poly-cis-decaprenylcistransferase, undecaprenyl diphosphate synthase, uds. Engineered: yes
|
|
Source:
|
 |
Escherichia coli. Organism_taxid: 562. Strain: bos-12. Expressed in: escherichia coli bl21. Expression_system_taxid: 511693.
|
|
Biol. unit:
|
 |
Dimer (from
)
|
|
Resolution:
|
 |
|
2.35Å
|
R-factor:
|
0.173
|
R-free:
|
0.254
|
|
|
Authors:
|
 |
S.-Y.Chang,T.-P.Ko,A.P.-C.Chen,A.H.-J.Wang,P.-H.Liang
|
Key ref:

|
 |
S.Y.Chang
et al.
(2004).
Substrate binding mode and reaction mechanism of undecaprenyl pyrophosphate synthase deduced from crystallographic studies.
Protein Sci,
13,
971-978.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
24-Dec-03
|
Release date:
|
13-Jan-04
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P60472
(UPPS_ECOLI) -
Ditrans,polycis-undecaprenyl-diphosphate synthase ((2E,6E)-farnesyl-diphosphate specific) from Escherichia coli (strain K12)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
253 a.a.
227 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.5.1.31
- ditrans,polycis-undecaprenyl-diphosphate synthase [(2E,6E)-farnesyl-
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Polyprenol biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
8 isopentenyl diphosphate + (2E,6E)-farnesyl diphosphate = di-trans,octa- cis-undecaprenyl diphosphate + 8 diphosphate
|
 |
 |
 |
 |
 |
8
×
isopentenyl diphosphate
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 (2E,6E)-farnesyl diphosphate
(2E,6E)-farnesyl diphosphate
|
=
|
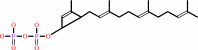 di-trans,octa- cis-undecaprenyl diphosphate
di-trans,octa- cis-undecaprenyl diphosphate
|
+
|
 8
×
diphosphate
8
×
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Protein Sci
13:971-978
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
Substrate binding mode and reaction mechanism of undecaprenyl pyrophosphate synthase deduced from crystallographic studies.
|
|
S.Y.Chang,
T.P.Ko,
A.P.Chen,
A.H.Wang,
P.H.Liang.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Undecaprenyl pyrophosphate synthase (UPPs) catalyzes eight consecutive
condensation reactions of farnesyl pyrophosphate (FPP) with isopentenyl
pyrophosphate (IPP) to form a 55-carbon long-chain product. We previously
reported the crystal structure of the apo-enzyme from Escherichia coli and the
structure of UPPs in complex with sulfate ions (resembling pyrophosphate of
substrate), Mg(2+), and two Triton molecules (product-like). In the present
study, FPP substrate was soaked into the UPPs crystals, and the complex
structure was solved. Based on the crystal structure, the pyrophosphate head
group of FPP is bound to the backbone NHs of Gly29 and Arg30 as well as the side
chains of Asn28, Arg30, and Arg39 through hydrogen bonds. His43 is close to the
C2 carbon of FPP and may stabilize the farnesyl cation intermediate during
catalysis. The hydrocarbon moiety of FPP is bound with hydrophobic amino acids
including Leu85, Leu88, and Phe89, located on the alpha3 helix. The binding mode
of FPP in cis-type UPPs is apparently different from that of trans-type and many
other prenyltransferases which utilize Asprich motifs for substrate binding via
Mg(2+). The new structure provides a plausible mechanism for the catalysis of
UPPs.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Figure 2. Interactions between FPP pyrophosphate head group
and the nearby amino acids in the active side. In A, the
pyrophosphate of FPP (F1) is hydrogen-bound to the backbone NH
and side chain of R30, and backbone NH of G29 as well as the
side chains of R39 and N28. Oxygen and nitrogen atoms are shown
as red and blue dots, respectively. All of the distances of
possible hydrogen bonds are indicated in Å, shown with red
dotted lines. In B, side chain of D26 forms hydrogen bonds with
the backbone NH of Gly27 and the side chain of R194 with the
distances indicated in Å. Moreover, R194 interacts with R200,
which is hydrogen-bound to E198. The segments of UPPs containing
residues 23-43 and 192-205 are represented by the red ribbon.
(C) Superimposition of the  A strand and
three A strand and
three  -helices ( -helices (  1, 1,  2, and 2, and  3) in the
active-site area from the closed and open conformations of UPPs.
The 3) in the
active-site area from the closed and open conformations of UPPs.
The  2 helix is
shown in red in the closed conformer and purple in the open
conformer. Several amino acids including L85, L88, F89, and W91
on this helix become closer to the bound FPP compared to their
positions (L85', L88', F89', and W91') in the open form. In D,
the Mg2+ (shown in yellow) near IPP is coordinated with H199
from A subunit, E213 from B subunit, and four waters. The
hydrocarbon parts of FPP and hypothetical IPP are represented by
ball-and-stick in yellow and black, respectively. The oxygen and
phosphate atoms in the pyrophosphate moiety are shown in red and
purple, respectively. The segments of A subunit of UPPs 23-43
and 192-205 are represented by red ribbon, and segments of B
subunit of apo-UPPs 210-215 by cyan ribbon. 2 helix is
shown in red in the closed conformer and purple in the open
conformer. Several amino acids including L85, L88, F89, and W91
on this helix become closer to the bound FPP compared to their
positions (L85', L88', F89', and W91') in the open form. In D,
the Mg2+ (shown in yellow) near IPP is coordinated with H199
from A subunit, E213 from B subunit, and four waters. The
hydrocarbon parts of FPP and hypothetical IPP are represented by
ball-and-stick in yellow and black, respectively. The oxygen and
phosphate atoms in the pyrophosphate moiety are shown in red and
purple, respectively. The segments of A subunit of UPPs 23-43
and 192-205 are represented by red ribbon, and segments of B
subunit of apo-UPPs 210-215 by cyan ribbon.
|
 |
Figure 4.
Figure 4. The proposed reaction mechanism of UPPs. Based on
the present crystal structure, Asp26 serves as a general base to
subtract a proton from IPP. This essential active-site amino
acid is near the proton at C2 carbon of IPP and ready to remove
it. The remaining electrons after deprotonation shift to form a
cis-double bond, and the carbanion intermediate attacks the C1
carbocation of FPP to form a condensation product. Eight cycles
total of IPP condensation generate the UPP product.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the Protein Society:
Protein Sci
(2004,
13,
971-978)
copyright 2004.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
B.Zhao,
C.Chun,
Z.Liu,
M.A.Horswill,
K.Pramanik,
G.A.Wilkinson,
R.Ramchandran,
and
R.Q.Miao
(2010).
Nogo-B receptor is essential for angiogenesis in zebrafish via Akt pathway.
|
| |
Blood,
116,
5423-5433.
|
 |
|
|
|
|
 |
L.V.Lee,
B.Granda,
K.Dean,
J.Tao,
E.Liu,
R.Zhang,
S.Peukert,
S.Wattanasin,
X.Xie,
N.S.Ryder,
R.Tommasi,
and
G.Deng
(2010).
Biophysical investigation of the mode of inhibition of tetramic acids, the allosteric inhibitors of undecaprenyl pyrophosphate synthase.
|
| |
Biochemistry,
49,
5366-5376.
|
 |
|
|
|
|
 |
A.Bouhss,
A.E.Trunkfield,
T.D.Bugg,
and
D.Mengin-Lecreulx
(2008).
The biosynthesis of peptidoglycan lipid-linked intermediates.
|
| |
FEMS Microbiol Rev,
32,
208-233.
|
 |
|
|
|
|
 |
C.J.Kuo,
R.T.Guo,
I.L.Lu,
H.G.Liu,
S.Y.Wu,
T.P.Ko,
A.H.Wang,
and
P.H.Liang
(2008).
Structure-based inhibitors exhibit differential activities against Helicobacter pylori and Escherichia coli undecaprenyl pyrophosphate synthases.
|
| |
J Biomed Biotechnol,
2008,
841312.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.Fujikura,
Y.Maki,
N.Ohya,
M.Satoh,
and
T.Koyama
(2008).
Kinetic studies of Micrococcus luteus B-P 26 undecaprenyl diphosphate synthase reaction using 3-desmethyl allylic substrate analogs.
|
| |
Biosci Biotechnol Biochem,
72,
851-855.
|
 |
|
|
|
|
 |
L.N.Kinch,
K.Ginalski,
and
N.V.Grishin
(2006).
Site-2 protease regulated intramembrane proteolysis: sequence homologs suggest an ancient signaling cascade.
|
| |
Protein Sci,
15,
84-93.
|
 |
|
|
|
|
 |
Y.Kharel,
S.Takahashi,
S.Yamashita,
and
T.Koyama
(2006).
Manipulation of prenyl chain length determination mechanism of cis-prenyltransferases.
|
| |
FEBS J,
273,
647-657.
|
 |
|
|
|
|
 |
F.Bouvier,
A.Rahier,
and
B.Camara
(2005).
Biogenesis, molecular regulation and function of plant isoprenoids.
|
| |
Prog Lipid Res,
44,
357-429.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

