 |
PDBsum entry 1adf

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase(NAD(a)-choh(d))
|
PDB id
|
|

|

|
1adf
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase(NAD(a)-choh(d))
|
 |
|
Title:
|
 |
Crystallographic studies of two alcohol dehydrogenase-bound analogs of thiazole-4-carboxamide adenine dinucleotide (tad), the active anabolite of the antitumor agent tiazofurin
|
|
Structure:
|
 |
Alcohol dehydrogenase. Chain: a. Engineered: yes
|
|
Source:
|
 |
Equus caballus. Horse. Organism_taxid: 9796
|
|
Biol. unit:
|
 |
 Dimer (from
)
Dimer (from
)
|
|
Resolution:
|
 |
|
|
Authors:
|
 |
H.Li,W.A.Hallows,J.S.Punzi,V.E.Marquez,H.L.Carrell,K.W.Pankiewicz, K.A.Watanabe,B.M.Goldstein
|
Key ref:

|
 |
H.Li
et al.
(1994).
Crystallographic studies of two alcohol dehydrogenase-bound analogues of thiazole-4-carboxamide adenine dinucleotide (TAD), the active anabolite of the antitumor agent tiazofurin.
Biochemistry,
33,
23-32.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
18-Oct-93
|
Release date:
|
31-Jan-94
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P00327
(ADH1E_HORSE) -
Alcohol dehydrogenase E chain from Equus caballus
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
375 a.a.
374 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.1.1
- alcohol dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
a primary alcohol + NAD+ = an aldehyde + NADH + H+
|
|
2.
|
a secondary alcohol + NAD+ = a ketone + NADH + H+
|
|
 |
 |
 |
 |
 |
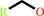 primary alcohol
primary alcohol
|
+
|
 NAD(+)
NAD(+)
|
=
|
 aldehyde
aldehyde
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
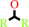 secondary alcohol
secondary alcohol
|
+
|
 NAD(+)
NAD(+)
|
=
|
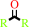 ketone
ketone
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Zn(2+) or Fe cation
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
33:23-32
(1994)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystallographic studies of two alcohol dehydrogenase-bound analogues of thiazole-4-carboxamide adenine dinucleotide (TAD), the active anabolite of the antitumor agent tiazofurin.
|
|
H.Li,
W.H.Hallows,
J.S.Punzi,
V.E.Marquez,
H.L.Carrell,
K.W.Pankiewicz,
K.A.Watanabe,
B.M.Goldstein.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Thiazole-4-carboxamide adenine dinucleotide (TAD) is the active anabolite of the
antitumor drug tiazofurin. Beta-methylene TAD (beta-TAD) is a
phosphodiesterase-resistant analogue of TAD, active in tiazofurin-resistant
cells. Beta-methylene SAD (beta-SAD) is the active selenium derivative of
beta-TAD. Both agents are analogues of the cofactor NAD and are capable of
acting as general dehydrogenase inhibitors. Crystal structures of beta-TAD and
beta-SAD bound to horse liver alcohol dehydrogenase (LADH) are presented at 2.9
and 2.7 A, respectively. Both complexes crystallize in the orthorhombic space
group C222(1) and are isomorphous to apo-LADH. Complexes containing beta-TAD and
beta-SAD were refined to crystallographic R values of 15% and 16%, respectively,
for reflections between 8 A and the minimum d spacing. Conformations of both
inhibitors are similar. beta-TAD and beta-SAD bind to the "open" form of LADH in
the normal cofactor-binding cleft between the coenzyme and catalytic domains of
each monomer. Binding at the adenosine end of each inhibitor resembles that of
NAD. However, the positions of the thiazole and selenazole heterocycles are
displaced away from the catalytic Zn cation by approximately 4 A. Close
intramolecular S-O and Se-O contacts observed in the parent nucleoside analogues
are maintained in both LADH-bound beta-TAD and beta-SAD, respectively. These
conformational constraints may influence the binding specificity of the
inhibitors.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
B.V.Plapp
(2010).
Conformational changes and catalysis by alcohol dehydrogenase.
|
| |
Arch Biochem Biophys,
493,
3.
|
 |
|
|
|
|
 |
A.Pennacchio,
L.Esposito,
A.Zagari,
M.Rossi,
and
C.A.Raia
(2009).
Role of Tryptophan 95 in substrate specificity and structural stability of Sulfolobus solfataricus alcohol dehydrogenase.
|
| |
Extremophiles,
13,
751-761.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
B.K.Sarma,
and
G.Mugesh
(2009).
Theoretical Investigation on the Effect of Different Nitrogen Donors on Intramolecular SeN Interactions.
|
| |
Chemphyschem,
10,
3013-3020.
|
 |
|
|
|
|
 |
L.Hedstrom
(2009).
IMP dehydrogenase: structure, mechanism, and inhibition.
|
| |
Chem Rev,
109,
2903-2928.
|
 |
|
|
|
|
 |
M.B.Stocks,
S.Hayward,
and
S.D.Laycock
(2009).
Interacting with the biomolecular solvent accessible surface via a haptic feedback device.
|
| |
BMC Struct Biol,
9,
69.
|
 |
|
|
|
|
 |
S.Hayward,
and
A.Kitao
(2006).
Molecular dynamics simulations of NAD+-induced domain closure in horse liver alcohol dehydrogenase.
|
| |
Biophys J,
91,
1823-1831.
|
 |
|
|
|
|
 |
R.Destro,
R.Soave,
M.Barzaghi,
and
L.Lo Presti
(2005).
Progress in the understanding of drug-receptor interactions, Part 1: experimental charge-density study of an angiotensin II receptor antagonist (C30H30N6O3S) at T = 17 K.
|
| |
Chemistry,
11,
4621-4634.
|
 |
|
|
|
|
 |
C.Brockmann,
A.Diehl,
K.Rehbein,
H.Strauss,
P.Schmieder,
B.Korn,
R.Kühne,
and
H.Oschkinat
(2004).
The oxidized subunit B8 from human complex I adopts a thioredoxin fold.
|
| |
Structure,
12,
1645-1654.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
B.M.Goldstein,
and
T.D.Colby
(2000).
Conformational constraints in NAD analogs: implications for dehydrogenase binding and specificity.
|
| |
Adv Enzyme Regul,
40,
405-426.
|
 |
|
|
|
|
 |
C.Schalk-Hihi,
and
G.D.Markham
(1999).
The conformations of a substrate and a product bound to the active site of S-adenosylmethionine synthetase.
|
| |
Biochemistry,
38,
2542-2550.
|
 |
|
|
|
|
 |
T.D.Colby,
K.Vanderveen,
M.D.Strickler,
G.D.Markham,
and
B.M.Goldstein
(1999).
Crystal structure of human type II inosine monophosphate dehydrogenase: implications for ligand binding and drug design.
|
| |
Proc Natl Acad Sci U S A,
96,
3531-3536.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.Schalk-Hihi,
Y.Z.Zhang,
and
G.D.Markham
(1998).
The conformation of NADH bound to inosine 5'-monophosphate dehydrogenase determined by transferred nuclear Overhauser effect spectroscopy.
|
| |
Biochemistry,
37,
7608-7616.
|
 |
|
|
|
|
 |
T.D.Colby,
B.J.Bahnson,
J.K.Chin,
J.P.Klinman,
and
B.M.Goldstein
(1998).
Active site modifications in a double mutant of liver alcohol dehydrogenase: structural studies of two enzyme-ligand complexes.
|
| |
Biochemistry,
37,
9295-9304.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
K.W.Pankiewicz
(1997).
Novel nicotinamide adenine dinucleotide analogues as potential anticancer agents: quest for specific inhibition of inosine monophosphate dehydrogenase.
|
| |
Pharmacol Ther,
76,
89.
|
 |
|
|
|
|
 |
L.Hemmingsen,
R.Bauer,
M.J.Bjerrum,
H.W.Adolph,
M.Zeppezauer,
and
E.Cedergren-Zeppezauer
(1996).
The protein conformation of Cd-substituted horse liver alcohol dehydrogenase and its metal-site coordination geometry in binary and ternary inhibitor complexes.
|
| |
Eur J Biochem,
241,
546-551.
|
 |
|
|
|
|
 |
M.Li,
F.Dyda,
I.Benhar,
I.Pastan,
and
D.R.Davies
(1996).
Crystal structure of the catalytic domain of Pseudomonas exotoxin A complexed with a nicotinamide adenine dinucleotide analog: implications for the activation process and for ADP ribosylation.
|
| |
Proc Natl Acad Sci U S A,
93,
6902-6906.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

