 |
PDBsum entry 3i4c

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
3i4c
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Crystal structure of sulfolobus solfataricus adh(ssadh) double mutant (w95l,n249y)
|
|
Structure:
|
 |
NAD-dependent alcohol dehydrogenase. Chain: a, b, c, d, e, h. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Sulfolobus solfataricus. Organism_taxid: 2287. Gene: adh, sso2536. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
2.00Å
|
R-factor:
|
0.228
|
R-free:
|
0.250
|
|
|
Authors:
|
 |
L.Esposito,A.Pennacchio,A.Zagari,M.Rossi,C.A.Raia
|
|
Key ref:
|
 |
A.Pennacchio
et al.
(2009).
Role of tryptophan 95 in substrate specificity and structural stability of Sulfolobus solfataricus alcohol dehydrogenase.
Extremophiles,
13,
751-761.
PubMed id:
|
 |
|
Date:
|
 |
|
01-Jul-09
|
Release date:
|
21-Jul-09
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B, C, D, E, H:
E.C.1.1.1.1
- alcohol dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
a primary alcohol + NAD+ = an aldehyde + NADH + H+
|
|
2.
|
a secondary alcohol + NAD+ = a ketone + NADH + H+
|
|
 |
 |
 |
 |
 |
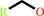 primary alcohol
primary alcohol
|
+
|
 NAD(+)
NAD(+)
|
=
|
 aldehyde
aldehyde
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
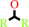 secondary alcohol
secondary alcohol
|
+
|
 NAD(+)
NAD(+)
|
=
|
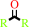 ketone
ketone
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Zn(2+) or Fe cation
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Extremophiles
13:751-761
(2009)
|
|
PubMed id:
|
|
|
|

|
| |
|
Role of tryptophan 95 in substrate specificity and structural stability of Sulfolobus solfataricus alcohol dehydrogenase.
|
|
A.Pennacchio,
L.Esposito,
A.Zagari,
M.Rossi,
C.A.Raia.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
A mutant of the thermostable NAD(+)-dependent (S)-stereospecific alcohol
dehydrogenase from Sulfolobus solfataricus (SsADH) which has a single
substitution, Trp95Leu, located at the substrate binding pocket, was fully
characterized to ascertain the role of Trp95 in discriminating between chiral
secondary alcohols suggested by the wild-type SsADH crystallographic structure.
The Trp95Leu mutant displays no apparent activity with short-chain primary and
secondary alcohols and poor activity with aromatic substrates and coenzyme.
Moreover, the Trp --> Leu substitution affects the structural stability of
the archaeal ADH, decreasing its thermal stability without relevant changes in
secondary structure. The double mutant Trp95Leu/Asn249Tyr was also purified to
assist in crystallographic analysis. This mutant exhibits higher activity but
decreased affinity toward aliphatic alcohols, aldehydes as well as NAD(+) and
NADH compared to the wild-type enzyme. The crystal structure of the
Trp95Leu/Asn249Tyr mutant apo form, determined at 2.0 A resolution, reveals a
large local rearrangement of the substrate site with dramatic consequences. The
Leu95 side-chain conformation points away from the catalytic metal center and
the widening of the substrate site is partially counteracted by a concomitant
change of Trp117 side chain conformation. Structural changes at the active site
are consistent with the reduced activity on substrates and decreased coenzyme
binding.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
G.A.Applegate,
R.W.Cheloha,
D.L.Nelson,
and
D.B.Berkowitz
(2011).
A new dehydrogenase from Clostridium acetobutylicum for asymmetric synthesis: dynamic reductive kinetic resolution entry into the Taxotère side chain.
|
| |
Chem Commun (Camb),
47,
2420-2422.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |
|

