D-arginine dehydrogenase
D-arginine dehydrogenase (DADH) is a flavin-dependent oxidoreductase. It is a member of the structural family consisting of D-amino acid oxidase, sarcosine oxidase, dimethyl glycine oxidase, and glycine oxidase. The enzyme has broad substrate specificity towards D-amino acids, particularly with cationic and hydrophobic D-amino acids. DADH and L-arginine dehydrogenase (LADH) make up a two-enzyme system involved in D- to L-arginine racemisation in pseudomonads and related species and both enzymes are products of the dauBAR operon, by D-arginine and D-lysine.
Crystallographic data shows that DADH from Pseudomonas aeruginosa (PaDADH) contains an active site loop L1 with two flexible segments that span the FAD-binding site and the substrate-binding site. It can occupy two distinct conformations, an open conformation competent for substrate binding and a closed conformation which is catalytically relevant and ligand-bound.
Reference Protein and Structure
- Sequence
-
Q9HXE3
 (1.4.99.6)
(1.4.99.6)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Pseudomonas aeruginosa PAO1 (Bacteria)

- PDB
-
3nye
- Crystal Structure of Pseudomonas aeruginosa D-Arginine Dehydrogenase in Complex with Imino-Arginine
(1.3 Å)



- Catalytic CATH Domains
-
3.30.9.10
 3.50.50.60
3.50.50.60  (see all for 3nye)
(see all for 3nye)
Enzyme Reaction (EC:1.4.99.6)
Enzyme Mechanism
Introduction
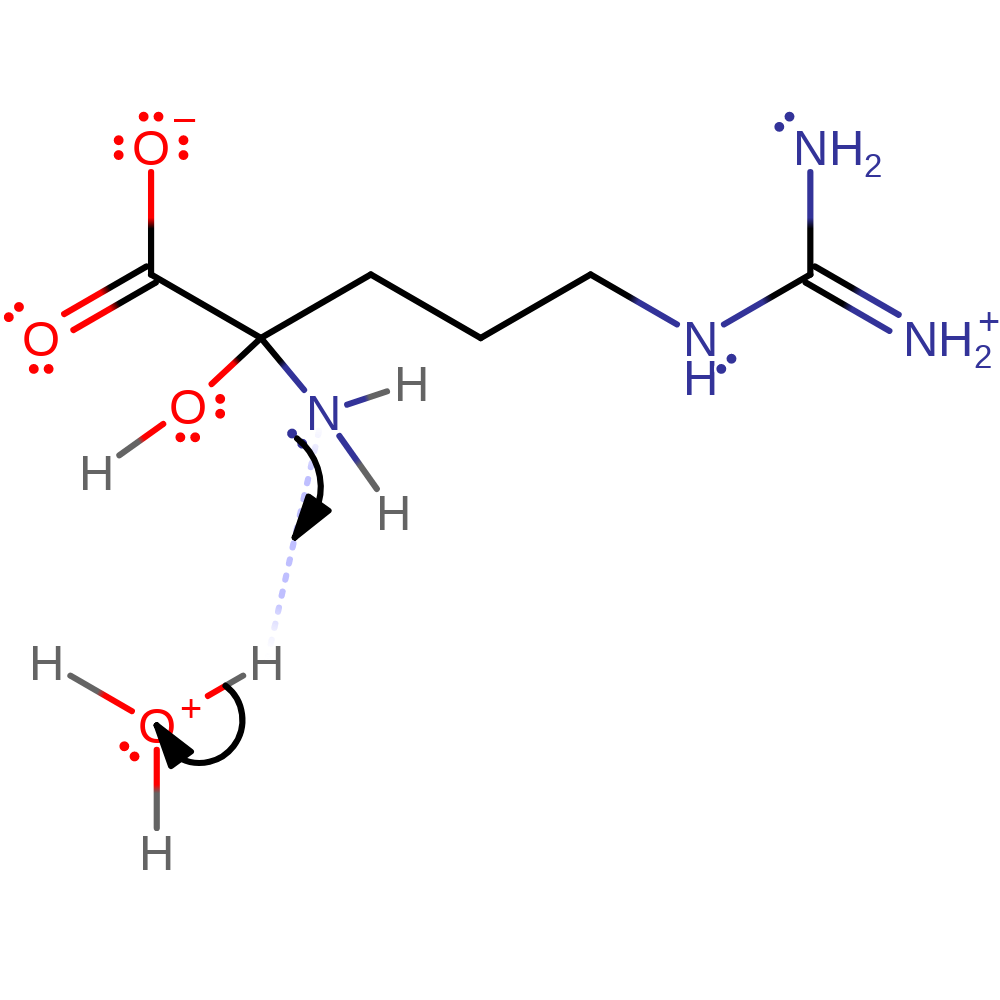
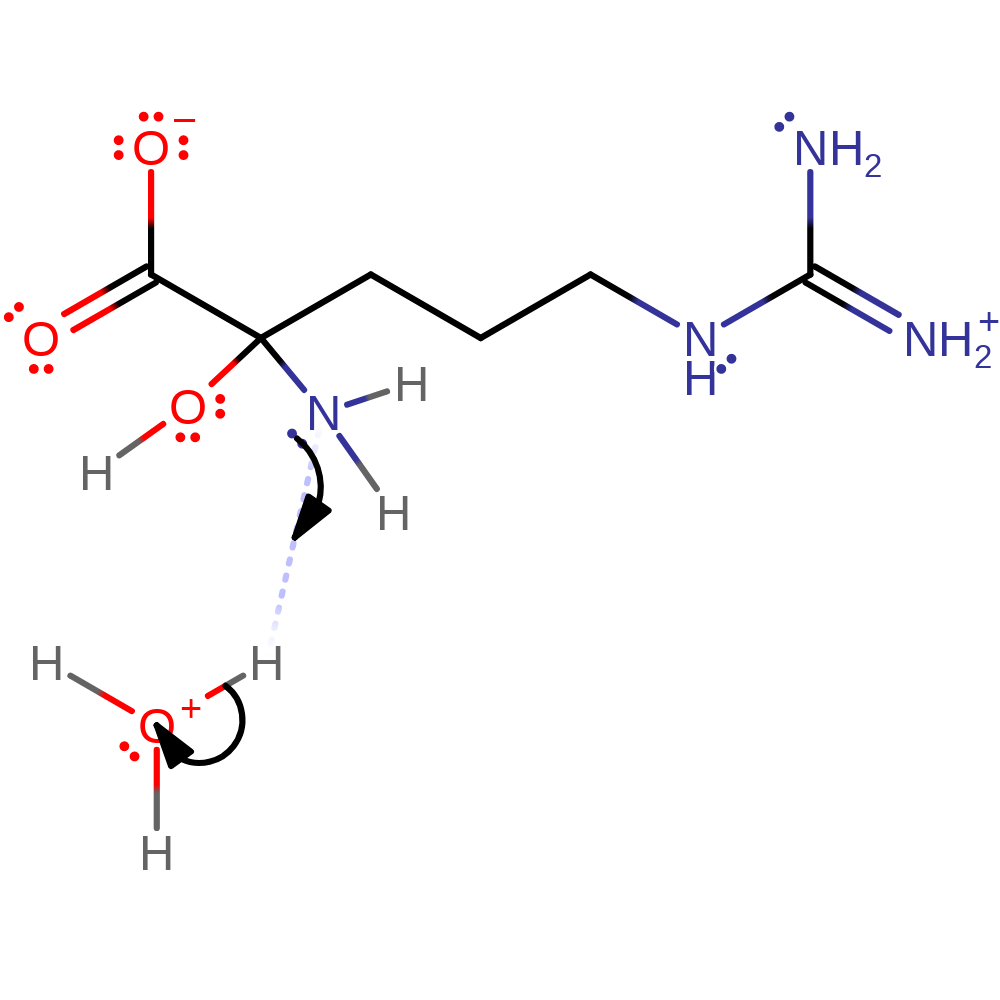
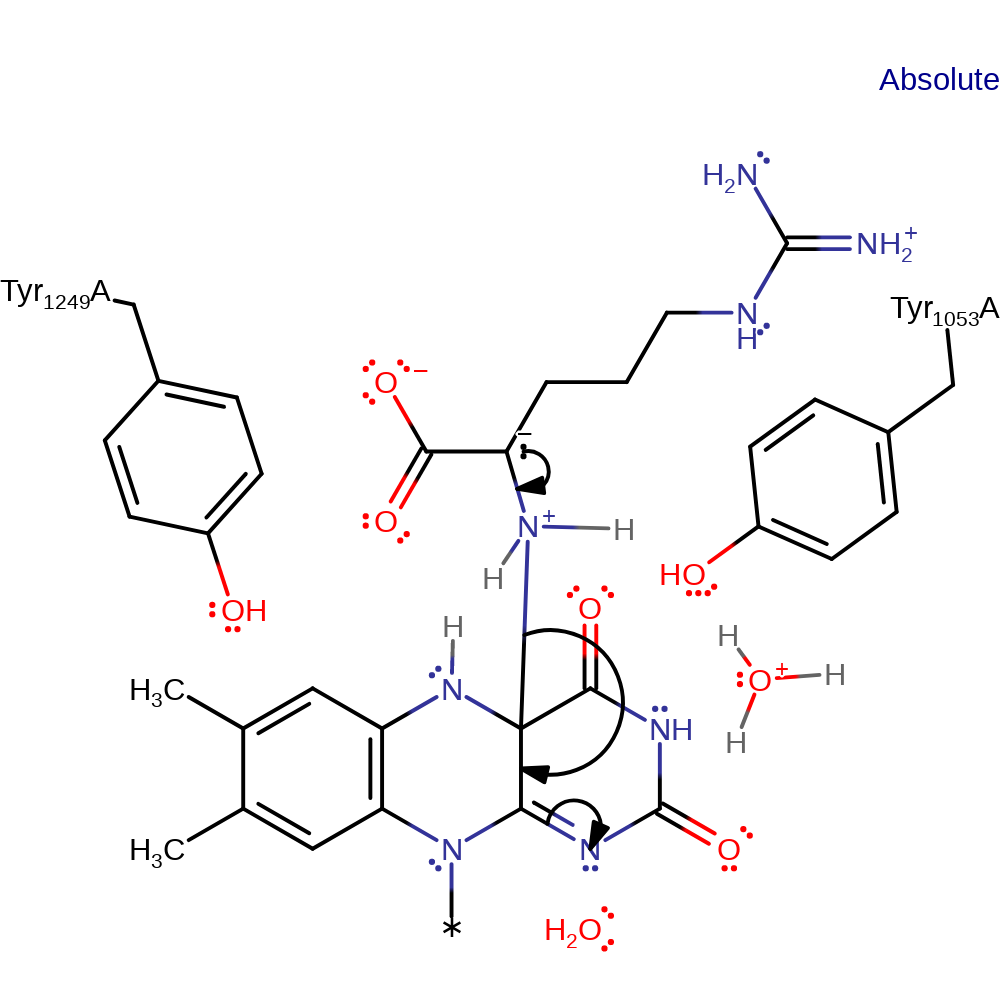
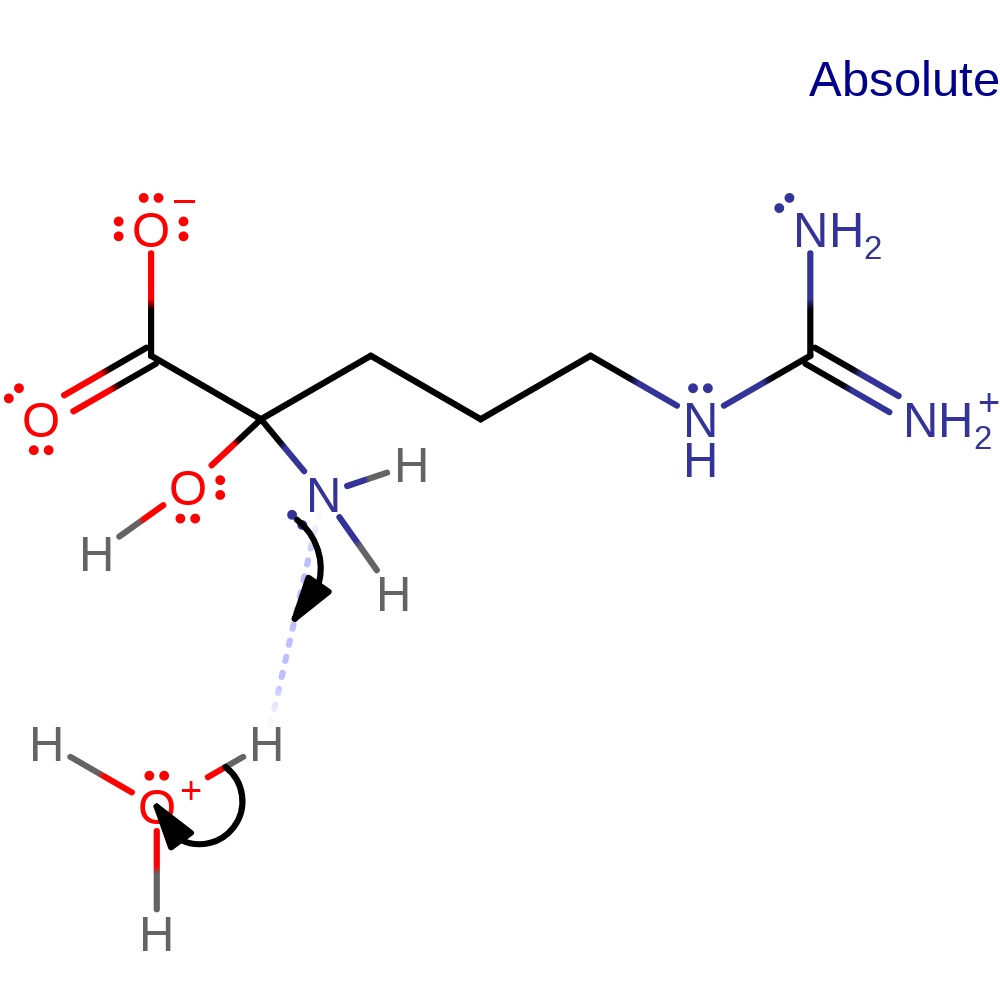
Hydride transfer mechanism supported by kinetic studies. In principle, transfer of an electron and a hydrogen atom from the substrate to the flavin is possible, but there is no evidence for presence of a radical as of yet. Deprotonation of the substrate α-amine occurs first, triggering CH bond cleavage through hydride transfer to the flavin N5 atom. Subsequent hydrolysis forms 5-guanidino-2-oxopentanoic acid and ammonia.
Catalytic Residues Roles
| UniProt | PDB* (3nye) | ||
| Tyr53, Tyr249 | Tyr1053(59)A, Tyr1249(255)A | Stabilise transition state in PaDADH to facilitate the hydride transfer mechanism. | transition state stabiliser |
| Glu87 | Glu1087(93)A | Stabilises D-arginine by engaging in an ionic interaction with the substrate guanido group. | electrostatic stabiliser |
| His48 | His1048(54)A | His48 forms a hydrogen-bond network with the amino group of the substrate, separated by two water molecules. The network is important to optimise substrate binding and prevent a slow proton release from the enzyme-substrate complex. | increase acidity |
| Ser45, Ala46 | Ser1045(51)A, Ala1046(52)A | In the FAD-binding site the side chains of S45 and A46 in loop L1, which do not interact directly with the substrate, adopt two alternate conformations depending on whether the ligand is present in the active site or not. | steric role |
Chemical Components
proton transfer, hydride transfer, cofactor used, intermediate formation, reaction occurs outside the enzyme, hydrolysis, bimolecular nucleophilic addition, intramolecular elimination, overall product formedReferences
- Gannavaram S et al. (2014), Biochemistry, 53, 6574-6583. Mechanistic and computational studies of the reductive half-reaction of tyrosine to phenylalanine active site variants of D-arginine dehydrogenase. DOI:10.1021/bi500917q. PMID:25243743.
- Ouedraogo D et al. (2017), Arch Biochem Biophys, 632, 192-201. Amine oxidation by d-arginine dehydrogenase in Pseudomonas aeruginosa. DOI:10.1016/j.abb.2017.06.013. PMID:28625766.
- Ball J et al. (2015), Arch Biochem Biophys, 568, 56-63. Importance of glutamate 87 and the substrate α-amine for the reaction catalyzed by D-arginine dehydrogenase. DOI:10.1016/j.abb.2015.01.017. PMID:25637657.
- Yuan H et al. (2011), J Am Chem Soc, 133, 18957-18965. Insights on the mechanism of amine oxidation catalyzed by D-arginine dehydrogenase through pH and kinetic isotope effects. DOI:10.1021/ja2082729. PMID:21999550.
- Fu G et al. (2011), Biochemistry, 50, 6292-6294. Atomic-resolution structure of an N5 flavin adduct in D-arginine dehydrogenase. DOI:10.1021/bi200831a. PMID:21707047.
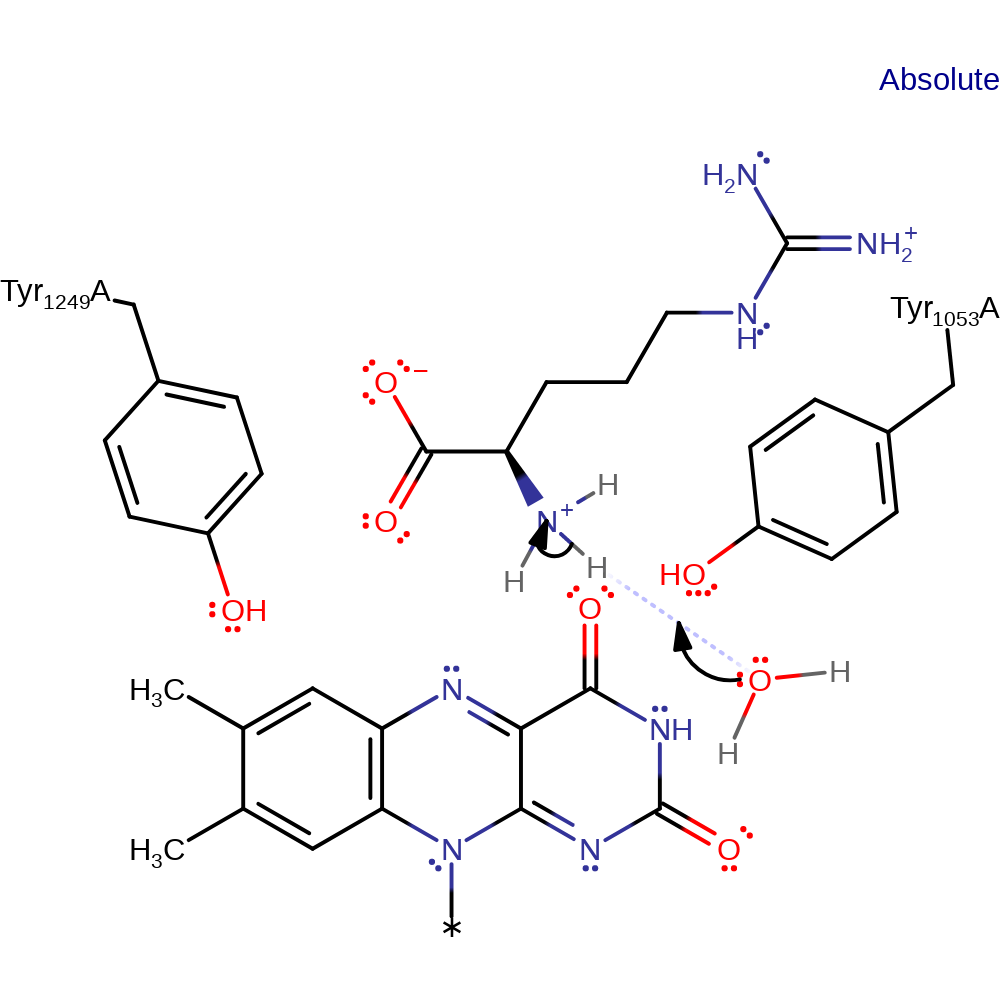
Step 1. Deprotonation of the substrate α-amine, triggering CH bond cleavage in the next step.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu1087(93)A | electrostatic stabiliser |
| His1048(54)A | increase acidity |
| Ser1045(51)A | steric role |
| Ala1046(52)A | steric role |
Chemical Components
proton transfer
Step 2. CH bond cleavage through the transfer of a hydride ion from the substrate to the flavin N5 atom.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr1053(59)A | transition state stabiliser |
| Tyr1249(255)A | transition state stabiliser |
| Glu1087(93)A | electrostatic stabiliser |
Chemical Components
hydride transfer, cofactor used, intermediate formation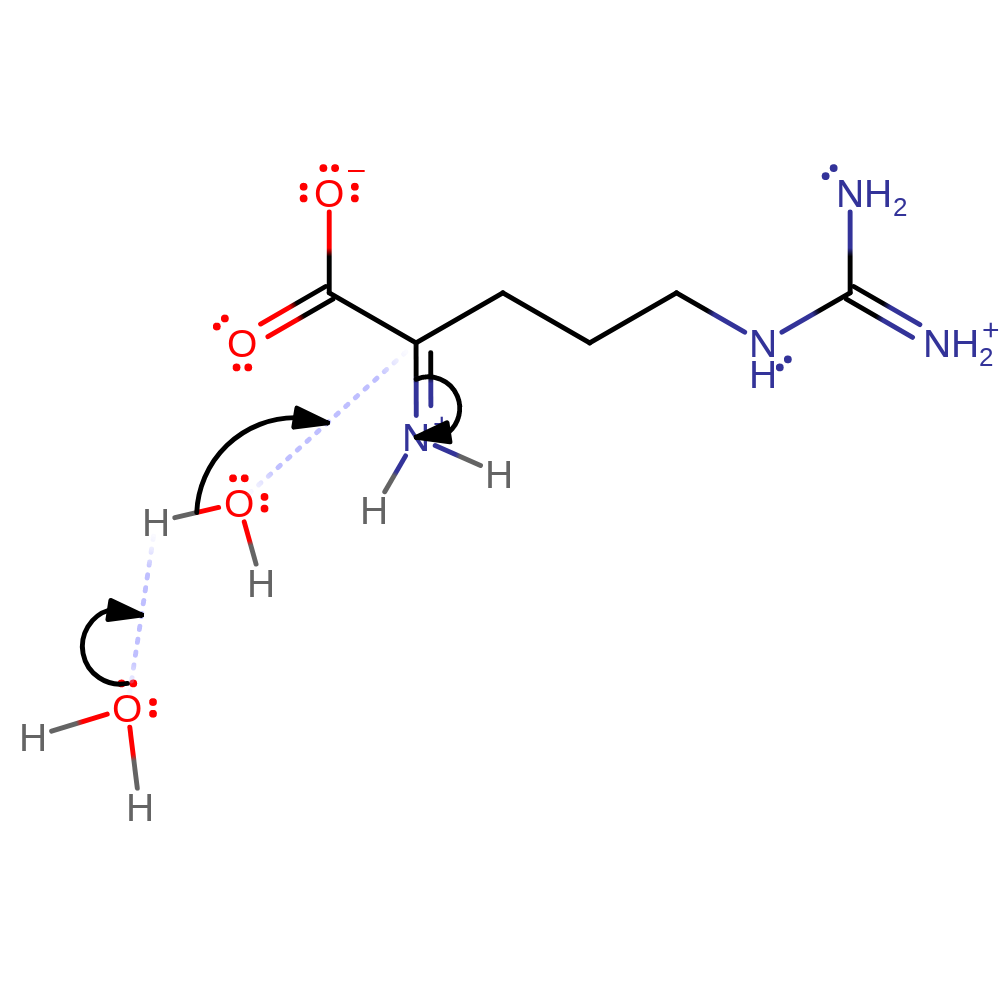
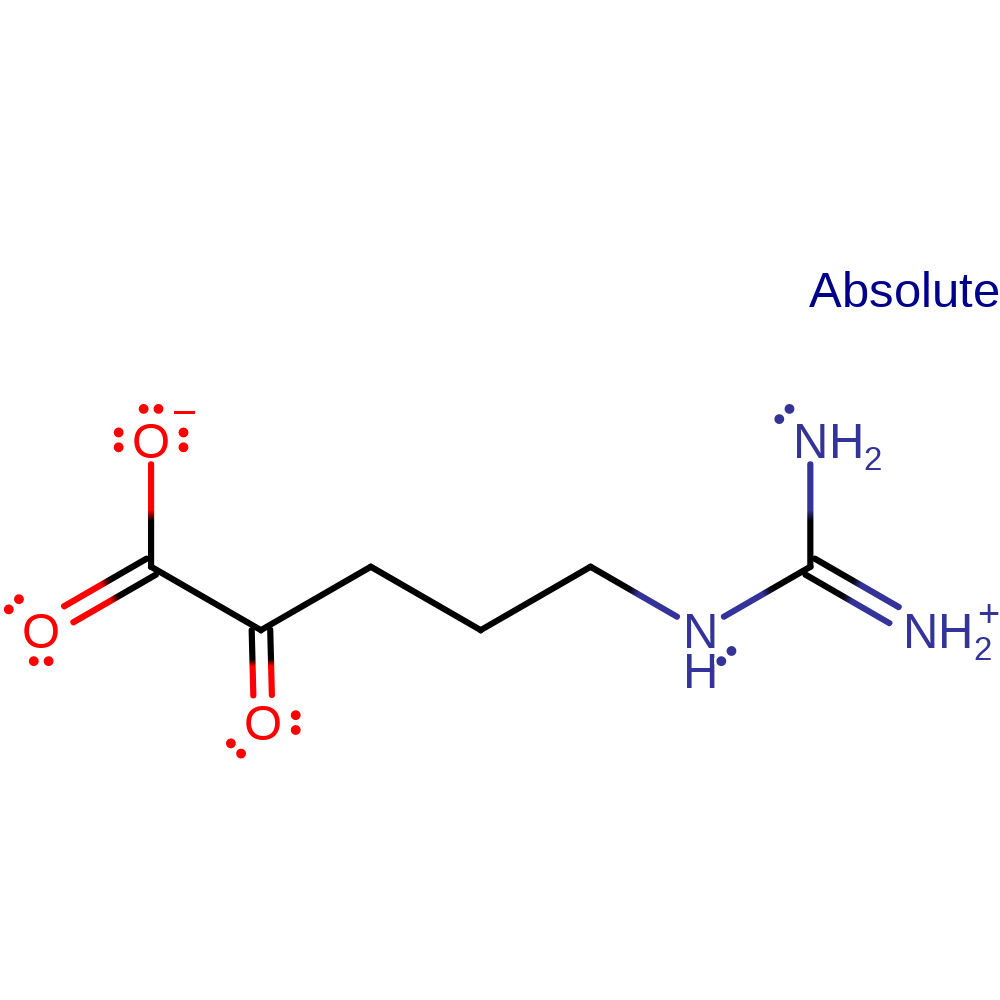
Step 3. Non-enzymatic hydrolysis of intermediate iminoacid to α-keto acid and ammonia. Begins with the addition of water.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
reaction occurs outside the enzyme, hydrolysis, ingold: bimolecular nucleophilic addition, proton transferCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
reaction occurs outside the enzyme, hydrolysis, proton transferCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
reaction occurs outside the enzyme, hydrolysis, ingold: intramolecular eliminationCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
reaction occurs outside the enzyme, hydrolysis, proton transfer, overall product formedIntroduction
A strong active site base abstracts the substrate α-proton with a pKa of ∼17. The resulting carbanion reacts with the flavin N5 atom, forming a covalent N5-flavin adduct. Cleavage of the substrate NH bond occurs stepwise with respect to CH bond cleavage, after formation of a covalent adduct with the flavin. Kinetic data suggesting NH and CH bond cleavage occur synchronously is not consistent with this mechanism.
Catalytic Residues Roles
| UniProt | PDB* (3nye) | ||
| Tyr53, Tyr249 | Tyr1053(59)A, Tyr1249(255)A | Stabilise transition state in PaDADH to facilitate the hydride transfer mechanism. | transition state stabiliser |
| Glu87 | Glu1087(93)A | Stabilises D-arginine by engaging in an ionic interaction with the substrate guanido group. | electrostatic stabiliser |
| His48 | His1048(54)A | His48 forms a hydrogen-bond network with the amino group of the substrate, separated by two water molecules. | increase acidity |
| Ser45, Ala46 | Ser1045(51)A, Ala1046(52)A | In the FAD-binding site the side chains of S45 and A46 in loop L1, which do not interact directly with the substrate, adopt two alternate conformations depending on whether the ligand is present in the active site or not. | steric role |
Chemical Components
inferred reaction step, proton transfer, bimolecular nucleophilic addition, aromatic unimolecular elimination by the conjugate base, native state of cofactor is not regenerated, intermediate formation, hydrolysis, reaction occurs outside the enzyme, intramolecular elimination, intermediate terminatedReferences
- Gannavaram S et al. (2014), Biochemistry, 53, 6574-6583. Mechanistic and computational studies of the reductive half-reaction of tyrosine to phenylalanine active site variants of D-arginine dehydrogenase. DOI:10.1021/bi500917q. PMID:25243743.
Catalytic Residues Roles
| Residue | Roles |
|---|---|
| Ser1045(51)A | steric role |
| Ala1046(52)A | steric role |
| Tyr1053(59)A | transition state stabiliser |
| Tyr1249(255)A | transition state stabiliser |
| His1048(54)A | increase acidity |
| Glu1087(93)A | electrostatic stabiliser |
Chemical Components
inferred reaction step, proton transfer
Step 2. Resulting carbanion reacts with the flavin N5 atom forming a covalent N5-flavin adduct.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
ingold: bimolecular nucleophilic addition
Step 3. Deprotonation of amine and cleavage of covalent N5-flavin adduct.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
ingold: aromatic unimolecular elimination by the conjugate base, native state of cofactor is not regenerated
Step 4. Non-enzymatic hydrolysis of intermediate iminoacid to α-keto acid and ammonia. Begins with the addition of water.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
intermediate formation, ingold: bimolecular nucleophilic addition, hydrolysis, reaction occurs outside the enzyme, proton transferCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
intermediate formation, proton transfer, hydrolysis, reaction occurs outside the enzymeCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
intermediate formation, ingold: intramolecular elimination, hydrolysis, reaction occurs outside the enzymeCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
intermediate terminated, proton transfer, hydrolysis, reaction occurs outside the enzymeIntroduction
A polar nucleophilic attack of the lone pair of electrons of the substrate α-amine at the flavin C4a atom may form a covalent C4a-flavin adduct. Cleavage of the substrate NH bond would occur stepwise with respect to CH bond cleavage, before formation of a covalent adduct with the flavin. Kinetic data suggesting NH and CH bond cleavage occur synchronously is not consistent with this mechanism.
Catalytic Residues Roles
| UniProt | PDB* (3nye) | ||
| Tyr53, Tyr249 | Tyr1053(59)A, Tyr1249(255)A | Stabilise transition state in PaDADH to facilitate the hydride transfer mechanism. | transition state stabiliser |
| Glu87 | Glu1087(93)A | Stabilises D-arginine by engaging in an ionic interaction with the substrate guanido group. | electrostatic stabiliser |
| His48 | His1048(54)A | His48 forms a hydrogen-bond network with the amino group of the substrate, separated by two water molecules. | increase acidity |
| Ser45, Ala46 | Ser1045(51)A, Ala1046(52)A | In the FAD-binding site the side chains of S45 and A46 in loop L1, which do not interact directly with the substrate, adopt two alternate conformations depending on whether the ligand is present in the active site or not. | steric role |
Chemical Components
proton transfer, bimolecular nucleophilic addition, cofactor used, intermediate formation, native state of cofactor is not regenerated, unimolecular elimination by the conjugate base, reaction occurs outside the enzyme, hydrolysis, intramolecular elimination, intermediate terminatedReferences
- Gannavaram S et al. (2014), Biochemistry, 53, 6574-6583. Mechanistic and computational studies of the reductive half-reaction of tyrosine to phenylalanine active site variants of D-arginine dehydrogenase. DOI:10.1021/bi500917q. PMID:25243743.
Catalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu1087(93)A | electrostatic stabiliser |
| Ser1045(51)A | steric role |
| Ala1046(52)A | steric role |
| Tyr1053(59)A | transition state stabiliser |
| Tyr1249(255)A | transition state stabiliser |
| His1048(54)A | increase acidity |
Chemical Components
proton transfer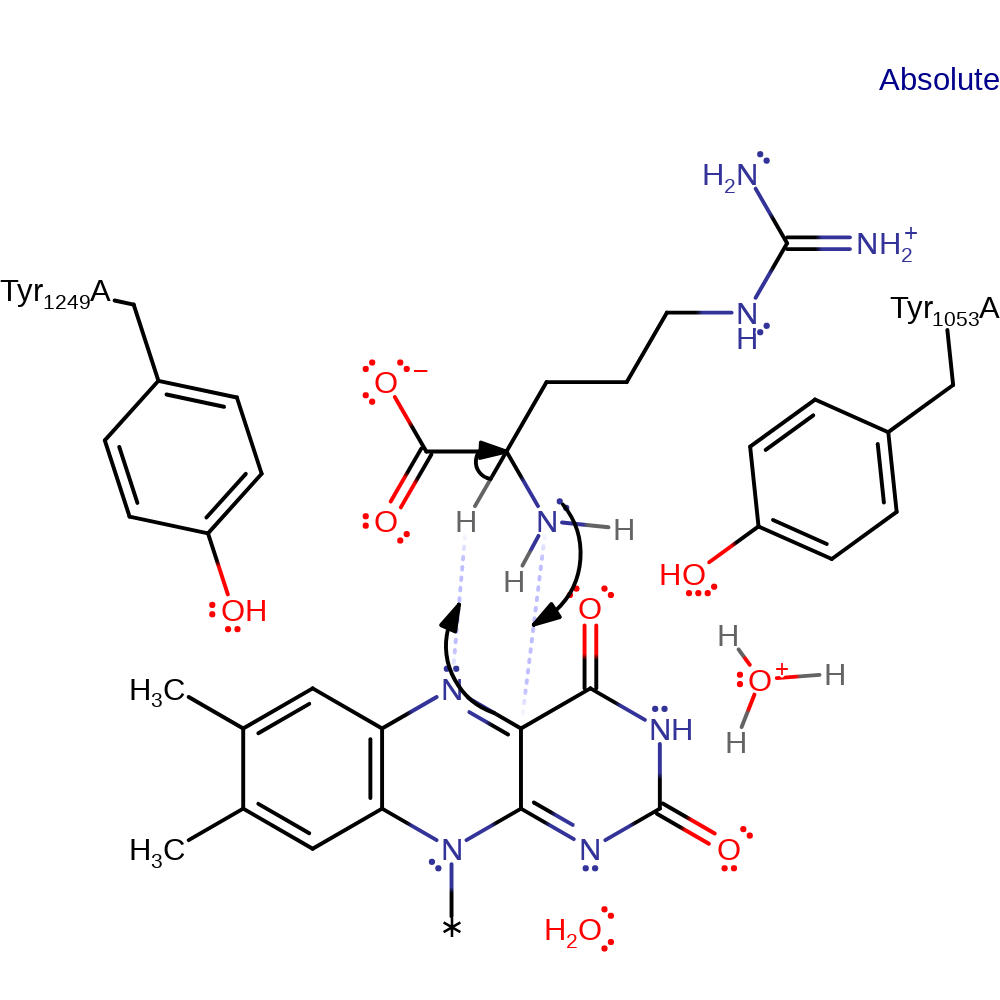
Step 2. Polar nucleophilic attack of the lone pair of electrons of the substrate α-amine at the flavin C4a atom forming a covalent C4a-flavin adduct.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
ingold: bimolecular nucleophilic addition, cofactor used, intermediate formationCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
native state of cofactor is not regenerated, ingold: unimolecular elimination by the conjugate base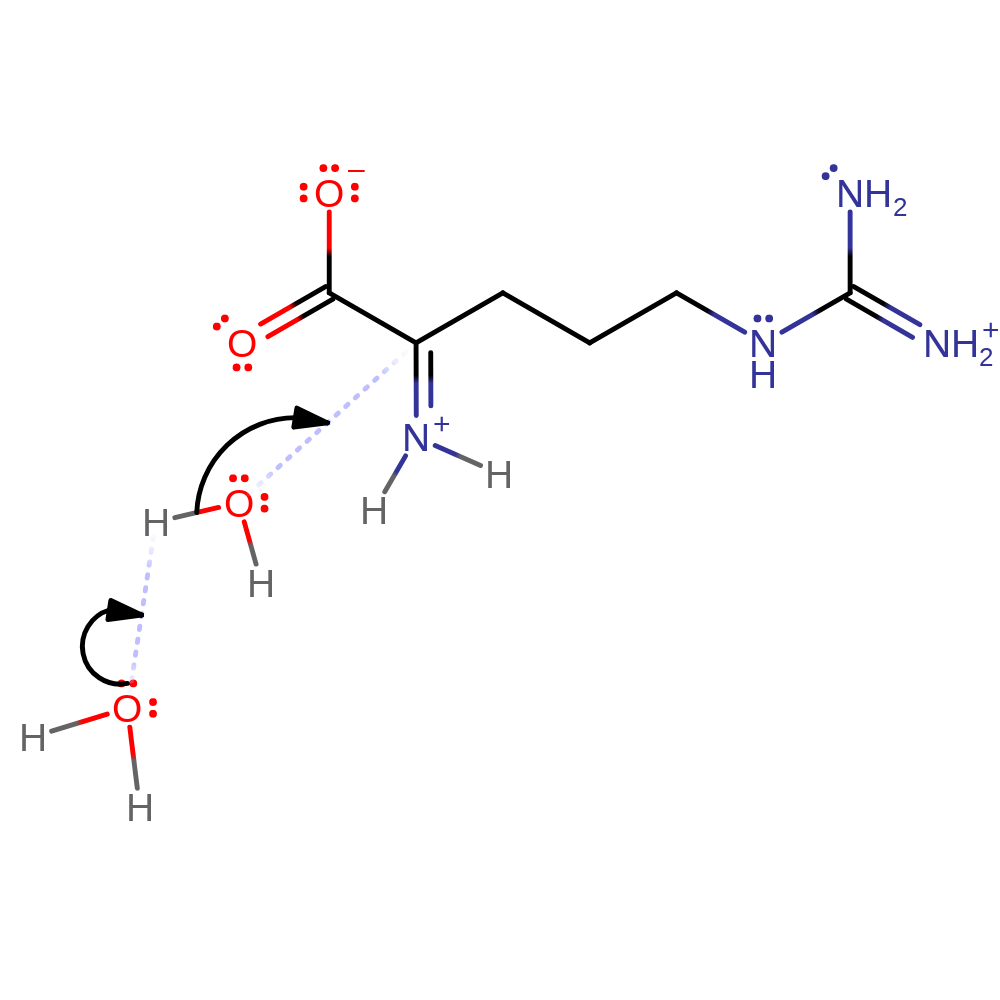
Step 4. Non-enzymatic hydrolysis of intermediate iminoacid to α-keto acid and ammonia. Begins with the addition of water.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
reaction occurs outside the enzyme, hydrolysis, ingold: bimolecular nucleophilic addition, intermediate formation, proton transferCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
reaction occurs outside the enzyme, hydrolysis, proton transfer, intermediate formationCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
reaction occurs outside the enzyme, hydrolysis, ingold: intramolecular elimination, intermediate formationCatalytic Residues Roles
| Residue | Roles |
|---|









 Download:
Download: 


 Download:
Download: 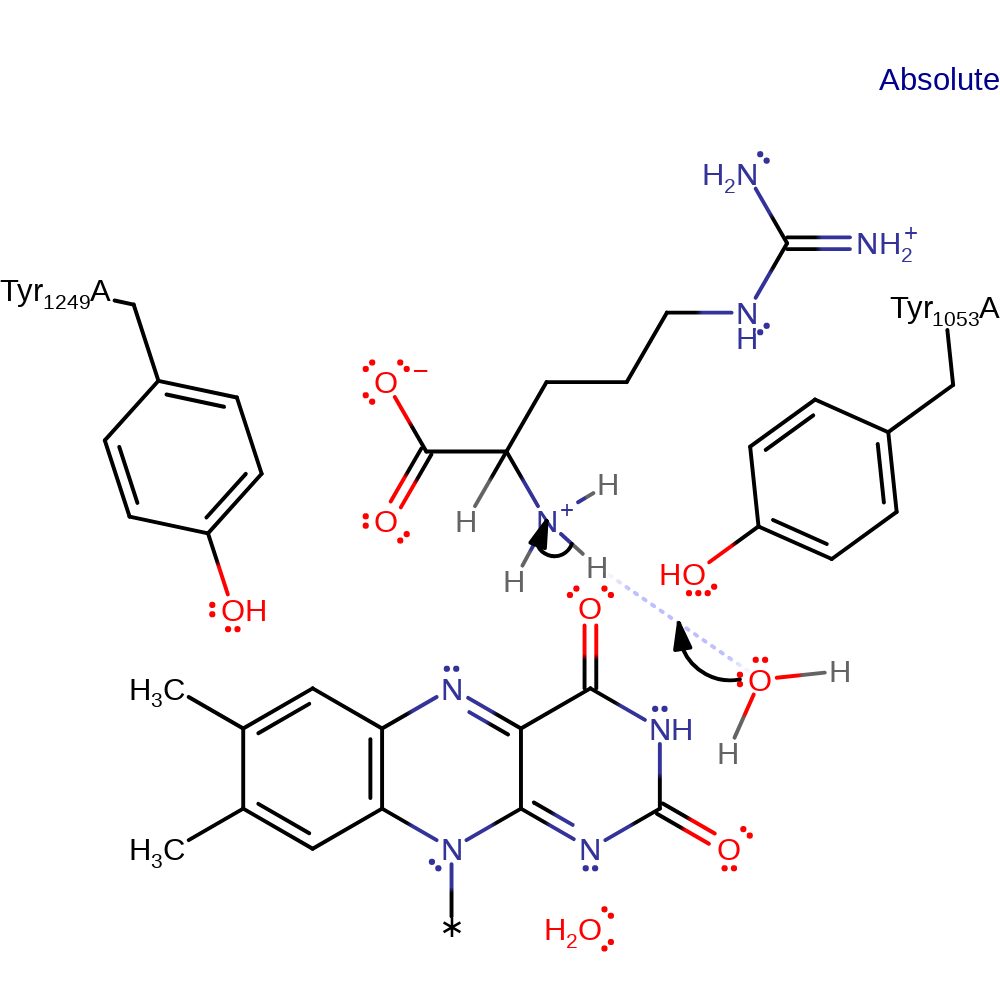


 Download:
Download: