Aldehyde oxidase
Aldehyde oxidase is a member of the xanthine oxidase family. It is a complex molybdoenzyme containing a Mo cofactor (Moco), flavin adenine nucleotide (FAD) and two Fe2S2 clusters. The enzyme has broad substrate specificity and catalyses a wide range of reactions. This includes the oxidation of aldehydes and several N-heterocycles, as well as the reduction of some substrates. In the human liver, the enzyme plays an essential role in the detoxification of xenobiotics and contributes to the hepatic clearance of many other drugs.
Reference Protein and Structure
- Sequence
-
Q06278
 (1.2.3.1, 1.17.3.-)
(1.2.3.1, 1.17.3.-)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Homo sapiens (Human)

- PDB
-
4uhx
- Human aldehyde oxidase in complex with phthalazine and thioridazine
(2.7 Å)



- Catalytic CATH Domains
- (see all for 4uhx)
- Cofactors
- Di-mu-sulfido-diiron(2+) (2)
Enzyme Reaction (EC:1.2.3.1)
Enzyme Mechanism
Introduction
The step-wise mechanism of oxidation of phthalazine (PHT) to phthalazin-1(2H)-one by hAOX1 begins with protonation of the substrate's N2 atom by Lys893. The hydroxyl group of molybdenum cofactor (Moco) subsequently attacks the substrate, and then a hydride transfer from the substrate to the sulphur atom of the Moco occurs. The rate-limiting step is the hydride transfer, determined by calculated free energy profiles.
Catalytic Residues Roles
| UniProt | PDB* (4uhx) | ||
| Glu1270 | Glu1270A | Deprotonates Moco, facilitating its nucleophilic attack on substrate. | electrostatic interaction, proton acceptor |
| Lys893 | Lys893A | Protonates substrate, activating it towards nucleophilic attack by Moco. | electrostatic interaction, proton donor |
Chemical Components
proton transfer, intermediate formation, bimolecular nucleophilic addition, hydride transfer, rate-determining step, electron transfer, intermediate collapse, cofactor used, redox reaction, inferred reaction step, radical formation, overall product formed, radical terminationReferences
- Ferreira P et al. (2020), ACS Catal, 10, 9276-9286. Catalytic Mechanism of Human Aldehyde Oxidase. DOI:10.1021/acscatal.0c02627.
- Mirzaei S et al. (2016), RSC Adv, 6, 109672-109680. Mechanistic study of allopurinol oxidation using aldehyde oxidase, xanthine oxidase and cytochrome P450 enzymes. DOI:10.1039/C6RA19197E.
- Coelho C et al. (2015),Human aldehyde oxidase in complex with phthalazine and thioridazine. DOI:10.2210/pdb4uhx/pdb.
- Cerqueira NM et al. (2015), J Biol Inorg Chem, 20, 209-217. Insights into the structural determinants of substrate specificity and activity in mouse aldehyde oxidases. DOI:10.1007/s00775-014-1198-2. PMID:25287365.
- Yamaguchi Y et al. (2007), J Biochem, 141, 513-524. Human xanthine oxidase changes its substrate specificity to aldehyde oxidase type upon mutation of amino acid residues in the active site: roles of active site residues in binding and activation of purine substrate. DOI:10.1093/jb/mvm053. PMID:17301077.
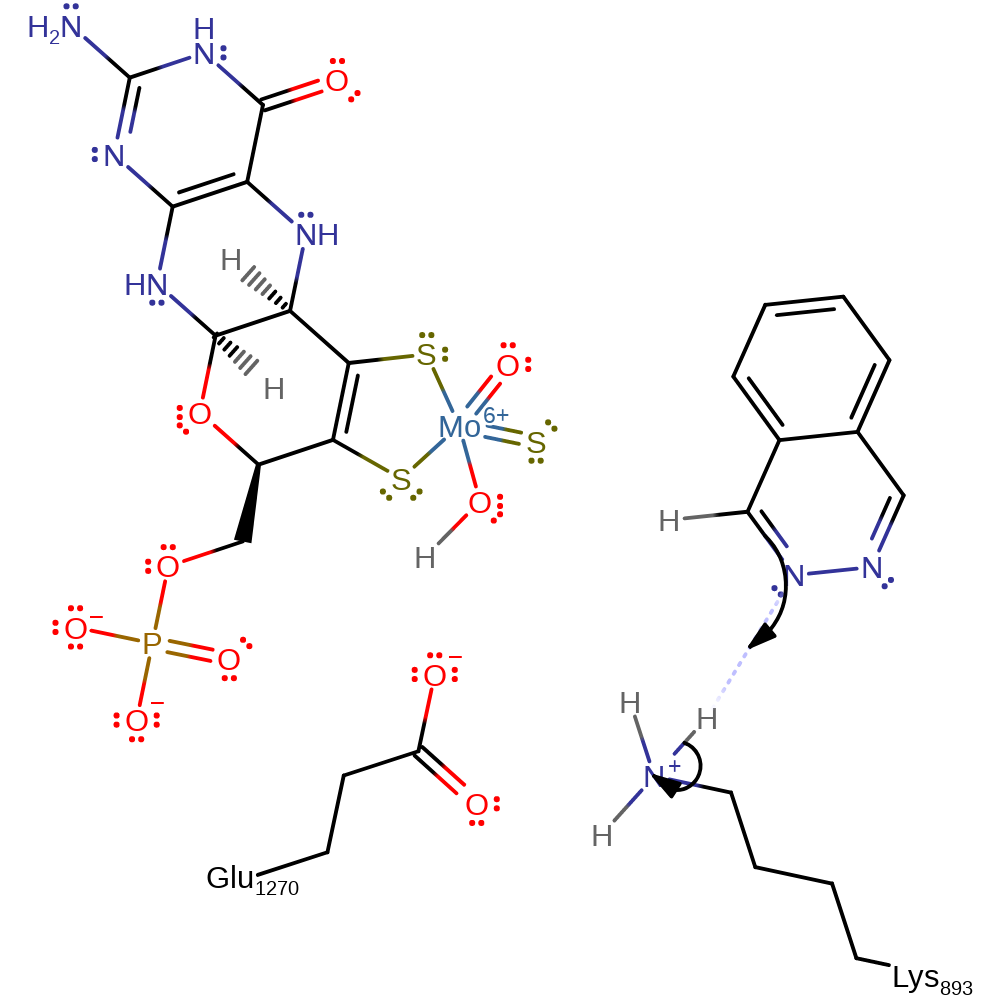
Step 1. Proton transfer from Lys893 to PHT. The transfer is barrierless and activates PHT for subsequent nucleophilic attack by the hydroxyl oxygen of the Moco (VI).
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys893A | proton donor |
| Glu1270A | electrostatic interaction |
Chemical Components
proton transfer
Step 2. Deprotonation of the hydroxyl group of the Moco (VI) by Glu1270 and nucleophilic attack on substrate, forming a tetrahedral intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys893A | electrostatic interaction |
| Glu1270A | proton acceptor |
Chemical Components
proton transfer, intermediate formation, ingold: bimolecular nucleophilic addition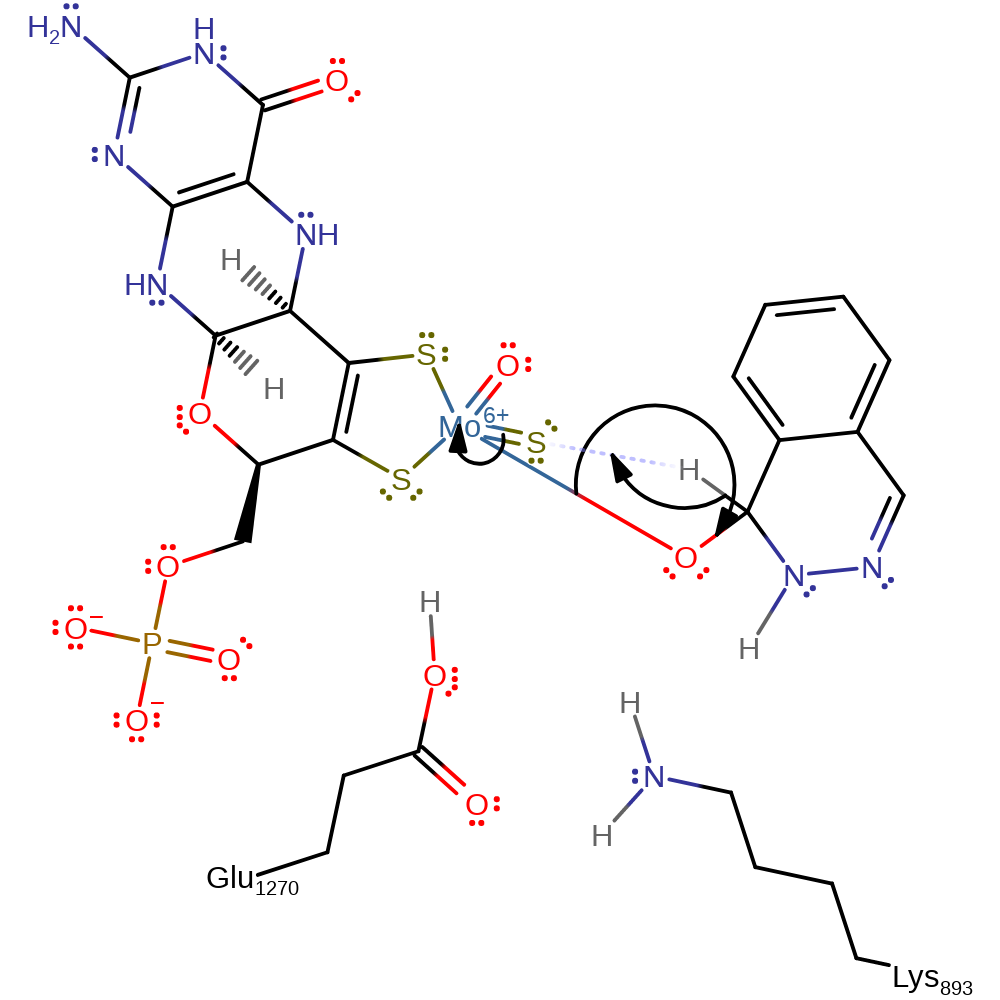
Step 3. Hydride transfer from substrate to the Moco (VI). This is the rate-limiting step of the reaction and reduces the Mo to (IV) .
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
hydride transfer, rate-determining step, electron transfer, intermediate collapse
Step 4. Single electron reduction and protonation of FAD by Moco (IV) forming Moco (V). Mechanism inferred by curator.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
cofactor used, redox reaction, electron transfer, proton transfer, inferred reaction step, radical formation
Step 5. Second single electron transfer and proton transfer from water forming FADH2 and oxidising Moco (V) to Moco (VI). Mechanism inferred by curator.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
proton transfer, overall product formed, cofactor used, inferred reaction step, electron transfer, radical termination, redox reactionIntroduction
Concerted mechanism. Moco-bound OH makes a nucleophilic attack on the carbon atom of the substrate, whilst the H atom is transferred to the S atom of the Moco. Supported by kinetic isotopic effects.
Catalytic Residues Roles
| UniProt | PDB* (4uhx) | ||
| Glu1270 | Glu1270A | Enhances stability of transition state and lowers activation energy of reaction. | electrostatic stabiliser |
| Lys893 | Lys893A | electrostatic stabiliser |
Chemical Components
bimolecular nucleophilic addition, hydride transfer, radical formation, inferred reaction step, proton transfer, electron transfer, redox reaction, cofactor used, radical termination, overall product formed, keto-enol tautomerisationReferences
- Montefiori M et al. (2017), ACS Omega, 2, 4237-4244. Aldehyde Oxidase: Reaction Mechanism and Prediction of Site of Metabolism. DOI:10.1021/acsomega.7b00658. PMID:30023718.
- Ferreira P et al. (2020), ACS Catal, 10, 9276-9286. Catalytic Mechanism of Human Aldehyde Oxidase. DOI:10.1021/acscatal.0c02627.
- Alfaro JF et al. (2008), J Org Chem, 73, 9469-9472. Studies on the mechanism of aldehyde oxidase and xanthine oxidase. DOI:10.1021/jo801053u. PMID:18998731.

Step 1. Moco-bound OH makes a nucleophilic attack on the carbon atom of the substrate, whilst hydride transfer occurs to the S atom of the Moco. Reduces Moco (VI) to Moco (IV).
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys893A | electrostatic stabiliser |
| Glu1270A | electrostatic stabiliser |
Chemical Components
ingold: bimolecular nucleophilic addition, hydride transfer
Step 2. Single electron reduction and protonation of FAD by Moco (IV) forming Moco (V). Mechanism inferred by curator.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
radical formation, inferred reaction step, proton transfer, electron transfer, redox reaction, cofactor used
Step 3. Second single electron transfer and proton transfer from water forming FADH2 and oxidising Moco (V) to Moco (VI). Mechanism inferred by curator.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
redox reaction, radical termination, electron transfer, inferred reaction step, cofactor used, overall product formed, proton transferCatalytic Residues Roles
| Residue | Roles |
|---|







 Download:
Download: 
 Download:
Download: