Mannonate dehydratase (uxuA)
Mannonate dehydratase (uxuA) converts D-mannonate to 2-dehydro-3-deoxy-D-gluconate. In contrast to the Enolase Superfamily equivalogue, this enzyme is dependent on a divalent iron cation. This protein is a member of the Xylose Isomerase-Like superfamily.
Reference Protein and Structure
- Sequence
-
P24215
 (4.2.1.8)
(4.2.1.8)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Escherichia coli K-12 (Bacteria)

- PDB
-
4eay
- Crystal structures of mannonate dehydratase from Escherichia coli strain K12 complexed with D-mannonate
(2.35 Å)



- Catalytic CATH Domains
-
3.20.20.150
 (see all for 4eay)
(see all for 4eay)
- Cofactors
- Iron(3+) (1)
Enzyme Reaction (EC:4.2.1.8)
Enzyme Mechanism
Introduction
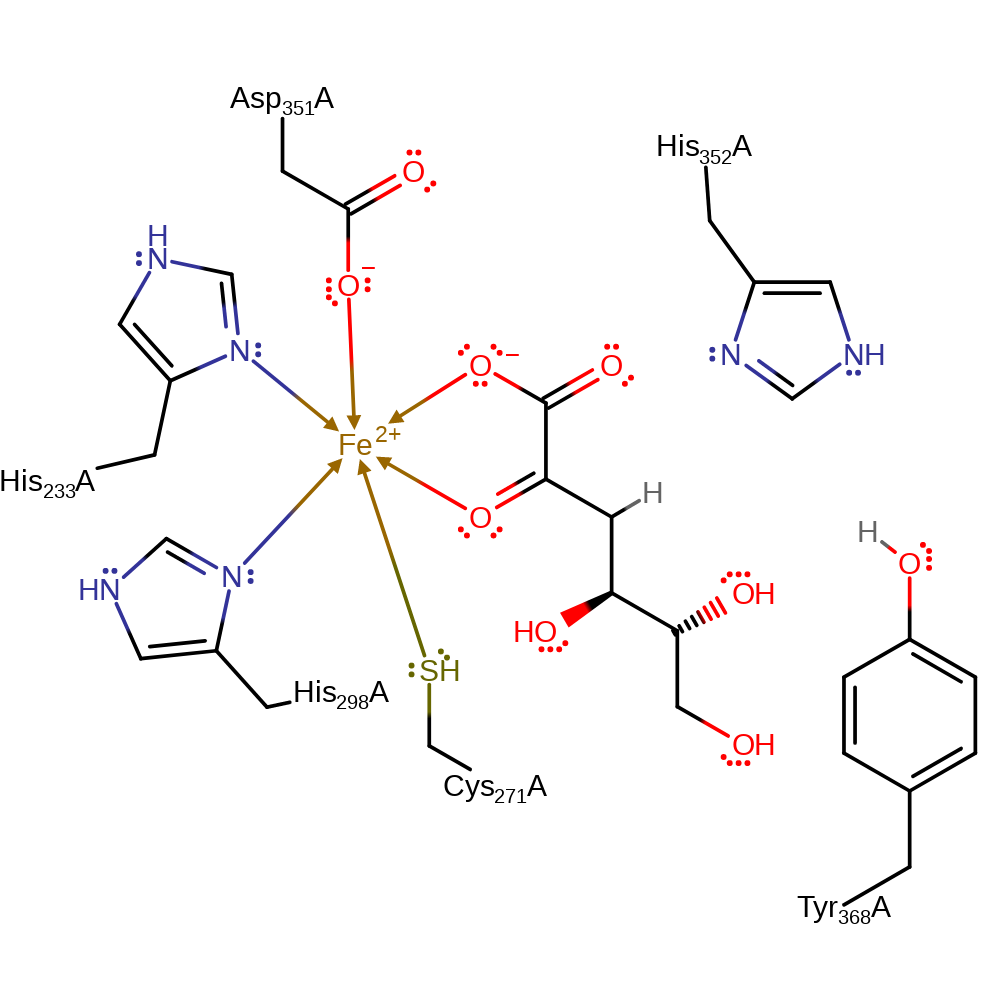
His311 abstracts the proton alpha to the carboxylate group (C2) to generate the stabilised enediolate intermediate. The vinylogous syn-elimination of the 3-OH is catalysed by Tyr368 and the following ketonisation with retention of configuration to generate the 2-keto-3-deoxy-D-gluconate product is probably catalysed by both Tyr368 and His311.
Catalytic Residues Roles
| UniProt | PDB* (4eay) | ||
| His352, Tyr368 | His352(372)A, Tyr368(388)A | Acts as a general acid/base. | proton acceptor, proton donor |
| His298, Asp351, Cys271, His233 | His298(318)A, Asp351(371)A, Cys271(291)A, His233(253)A | Forms part of the metal binding site. | metal ligand |
Chemical Components
proton transfer, overall reactant used, assisted keto-enol tautomerisation, dehydration, overall product formed, unimolecular elimination by the conjugate base, inferred reaction step, native state of enzyme regeneratedReferences
- Zhang Q et al. (2009), J Bacteriol, 191, 5832-5837. Crystal structures of Streptococcus suis mannonate dehydratase (ManD) and its complex with substrate: genetic and biochemical evidence for a catalytic mechanism. DOI:10.1128/JB.00599-09. PMID:19617363.
- Dreyer JL (1987), Eur J Biochem, 166, 623-630. The role of iron in the activation of mannonic and altronic acid hydratases, two Fe-requiring hydro-lyases. PMID:3038546.

Step 1. His311 abstracts a proton from the substrate in an assisted keto-enol tautomerisation.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp351(371)A | metal ligand |
| His233(253)A | metal ligand |
| Cys271(291)A | metal ligand |
| His298(318)A | metal ligand |
| His352(372)A | proton acceptor |
Chemical Components
proton transfer, overall reactant used, assisted keto-enol tautomerisationCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp351(371)A | metal ligand |
| His233(253)A | metal ligand |
| Cys271(291)A | metal ligand |
| His298(318)A | metal ligand |
| Tyr368(388)A | proton donor |
Chemical Components
dehydration, overall product formed, proton transfer, assisted keto-enol tautomerisation, ingold: unimolecular elimination by the conjugate base
Step 3. Inferred step in which the enol intermediate rearranges to form the 2-keto-3-deoxygluconate product and return the enzyme active site to its starting state.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp351(371)A | metal ligand |
| His233(253)A | metal ligand |
| Cys271(291)A | metal ligand |
| His298(318)A | metal ligand |
| His352(372)A | proton donor |
| Tyr368(388)A | proton acceptor |




 Download:
Download: