Mannonate dehydratase
Characterised enzymes in the mannonate dehydratase family (manD) catalyse the dehydration of mannonate to 2-dehydro-3-deoxy-D-gluconate, as part of the catabolism of D-glucuronate. Generally, genomes that encode a member of the mannonate dehydratase family do not have a UxuA (iron(II)-dependent mannoate dehydratase) homologue. Though UxuA also has the TIM barrel fold, it is not a member of the enolase superfamily. Thus, the enolase superfamily mannonate dehydratase family and the UxuAs are an example of convergent evolution of function.
Reference Protein and Structure
- Sequence
-
A4XF23
 (4.2.1.8)
(4.2.1.8)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Novosphingobium aromaticivorans DSM 12444 (Bacteria)

- PDB
-
2qjj
- Crystal structure of D-Mannonate dehydratase from Novosphingobium aromaticivorans
(1.8 Å)



- Catalytic CATH Domains
-
3.30.390.10
 3.20.20.120
3.20.20.120  (see all for 2qjj)
(see all for 2qjj)
- Cofactors
- Magnesium(2+) (1)
Enzyme Reaction (EC:4.2.1.8)
Enzyme Mechanism
Introduction
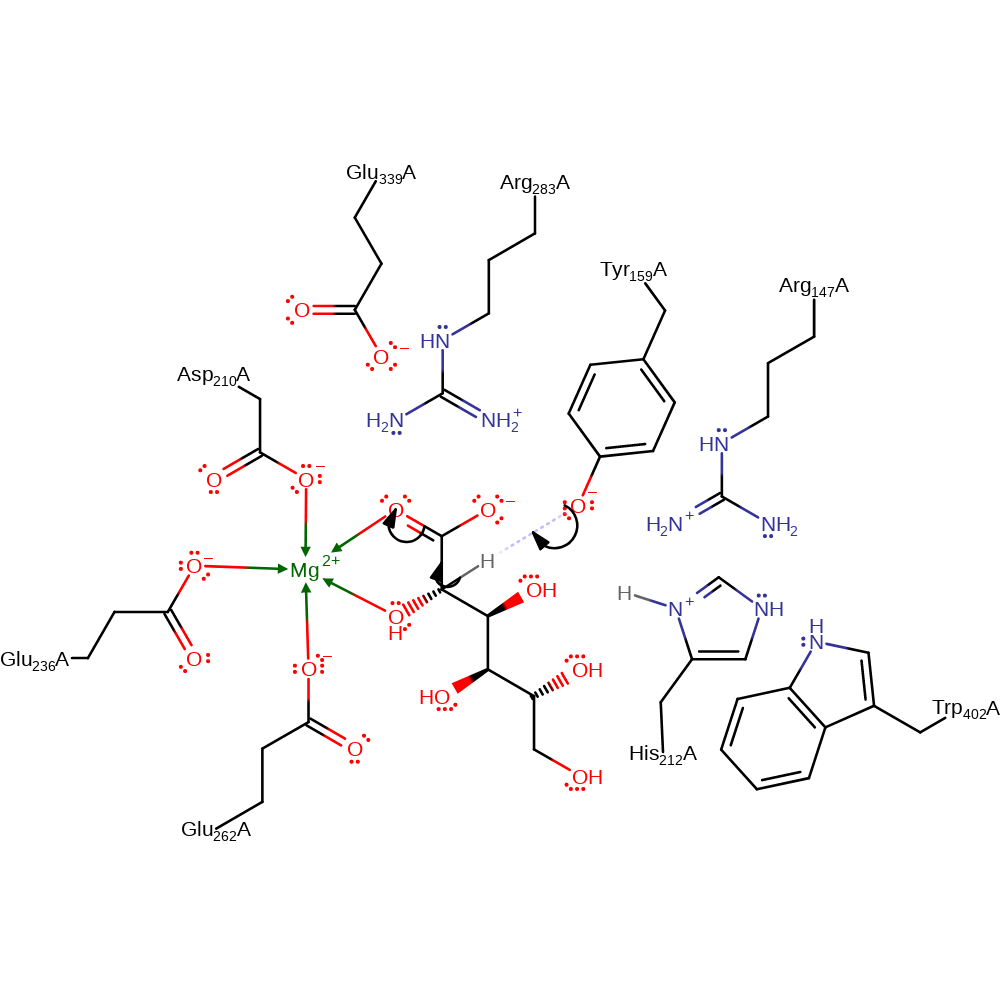
Tyr159, hydrogen-bonded to Arg147, is positioned to function as the base that abstracts the proton alpha to the carboxylate group (C2) to generate the stabilised enediolate intermediate. The vinylogous syn-elimination of the 3-OH followed by ketonisation with retention of configuration to generate the 2-keto-3-deoxy-D-gluconate product may be catalysed by Tyr159 and/or His212 (shown as His212 here).
Catalytic Residues Roles
| UniProt | PDB* (2qjj) | ||
| Arg147 | Arg147A | Modulates the pKa of the catalytic tyrosine residue through a hydrogen bonding network. | modifies pKa |
| Tyr159 | Tyr159A | Acts as a general acid/base. This residue is appropriately positioned to function as the base that abstracts the proton alpha to the carboxylate group. | proton acceptor, proton donor |
| His212 | His212A | Acts as a general acid/base. Thought to be the residue responsible for the protonation of the leaving hydroxyl group. | proton acceptor, proton donor |
| Asp210, Glu236, Glu262 | Asp210A, Glu236A, Glu262A | Forms part of the magnesium binding site. | metal ligand |
| Arg283, Glu339 | Arg283A, Glu339A | Help stabilise the reactive intermediates and transition states formed during the course of the reaction. | electrostatic stabiliser |
| Trp402 | Trp402A | Thought to help modulate the pKa of His212. | modifies pKa |
Chemical Components
proton transfer, assisted keto-enol tautomerisation, overall reactant used, overall product formed, native state of enzyme regeneratedReferences
- Rakus JF et al. (2007), Biochemistry, 46, 12896-12908. Evolution of enzymatic activities in the enolase superfamily: D-Mannonate dehydratase from Novosphingobium aromaticivorans. DOI:10.1021/bi701703w. PMID:17944491.
- Zhang Q et al. (2009), J Bacteriol, 191, 5832-5837. Crystal structures of Streptococcus suis mannonate dehydratase (ManD) and its complex with substrate: genetic and biochemical evidence for a catalytic mechanism. DOI:10.1128/JB.00599-09. PMID:19617363.
Catalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu339A | electrostatic stabiliser |
| Arg283A | electrostatic stabiliser |
| Trp402A | modifies pKa |
| Arg147A | modifies pKa |
| Glu236A | metal ligand |
| Glu262A | metal ligand |
| Asp210A | metal ligand |
| Tyr159A | proton acceptor |
Chemical Components
proton transfer, assisted keto-enol tautomerisation, overall reactant used
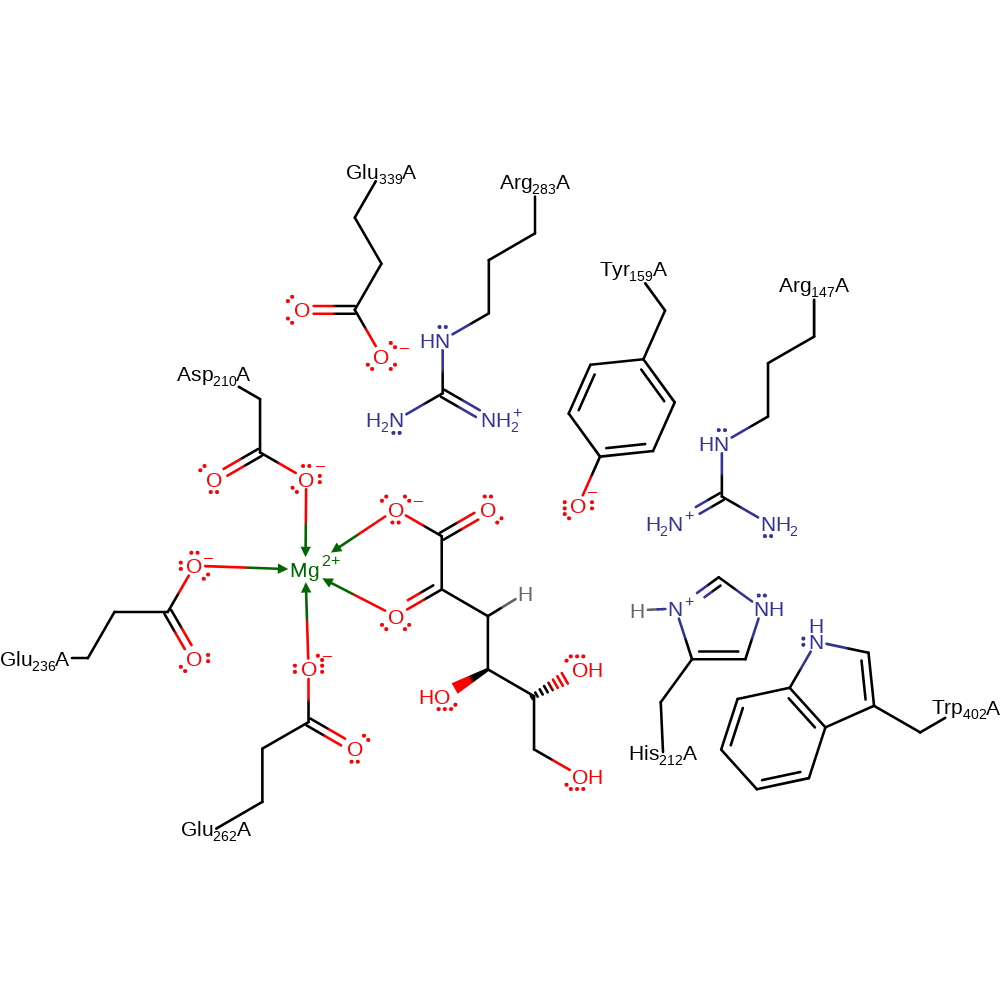
Step 2. The intermediate collapses, eliminating a water molecule with concomitant deprotonation of His212.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu339A | electrostatic stabiliser |
| Arg283A | electrostatic stabiliser |
| Trp402A | modifies pKa |
| Arg147A | modifies pKa |
| Glu236A | metal ligand |
| Glu262A | metal ligand |
| Asp210A | metal ligand |
| His212A | proton donor |
Chemical Components
assisted keto-enol tautomerisation, proton transfer, overall product formed
Step 3. His212 abstracts a proton from the hydroxyl coordinated to the Mg(II) ion in an assisted keto-enol tautomerisation. Tyr159 donates a proton to the intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg147A | modifies pKa |
| Trp402A | modifies pKa |
| Arg283A | electrostatic stabiliser |
| Glu339A | electrostatic stabiliser |
| Glu236A | metal ligand |
| Glu262A | metal ligand |
| Asp210A | metal ligand |
| Tyr159A | proton donor |
| His212A | proton acceptor |



 Download:
Download: