Methionine adenosyltransferase
In biological systems, methyl groups are transferred from a small number of donors to a large number of acceptors. S-adenosylmethionine (AdoMet) is the most widespread of these donors, and is synthesised solely by the action of AdoMet synthase.
The catalytic site of this enzyme, found in a cleft between two subunits, conducts an unusual reaction pathway where a triphosphate chain is cleaved from ATP as AdoMet is formed and the triphosphate is hydrolysed to diphosphate and phosphate before the product is released. There are three similar domains arranged around a pseudo-threefold symmetry axis.
Reference Protein and Structure
- Sequence
-
P31153
 (2.5.1.6)
(2.5.1.6)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Homo sapiens (Human)

- PDB
-
5a1i
- The structure of Humab MAT2A in complex with SAMe, Adenosine, Methionine and PPNP.
(1.09 Å)



- Catalytic CATH Domains
-
3.30.300.10
 (see all for 5a1i)
(see all for 5a1i)
- Cofactors
- Magnesium(2+) (2), Potassium(1+) (1) Metal MACiE
Enzyme Reaction (EC:2.5.1.6)
Enzyme Mechanism
Introduction
The reaction involves the cleavage of the triphosphate chain from ATP, in forming the product, and hydrolysis of the PPPi moiety to PPi and Pi before the AdoMet product is released.
Mechanistic studies have shown the AdoMet forming reaction to follow an SN2 mechanism, with the S atom of methionine attacking the C5 atom of ATP directly. His29 acts as a general acid, activated by the surrounding basic backbone amide groups, towards the O5' as the C5'-O5' bond cleaves. Simultaneously, the methionine sulphur attacks the developing cation. This reaction is followed by the hydrolysis of triphosphate to phosphate and pyrophosphate, providing energy for the removal of the reaction product from the active site.
The reaction requires divalent metal cations for activity, two binding sites have been identified both by structural information and EPR studies.
Catalytic Residues Roles
| UniProt | PDB* (5a1i) | ||
| His29 | His29A | Acts as a general acid/base by abstratcing donating a proton to the triphosphate leaving group. It returns to its initial protonation step through the abstraction of a proton from water. | proton acceptor, proton donor |
| Asp31 | Asp31A | Binds Mg(II) ion | metal ligand, electrostatic stabiliser |
| Lys32 (main-N), Asp31 | Lys32A (main-N), Asp31A | main chain amides help stabilise the anionic His29 | electrostatic stabiliser |
| Glu57, Asp258, Ala259 (main-C) | Glu57A, Asp258A, Ala259A (main-C) | Binds K(I) ion. | metal ligand |
| Lys181, Arg264, Lys265, Lys285 | Lys181A, Arg264A, Lys265A, Lys285A | Stabilises the negative charges on the triphosphate moiety. | electrostatic stabiliser |
| Phe250, Glu70 | Phe250A, Glu70A | Guides the steric outcome of the reaction. | steric role |
| Lys289, Asp291 | Lys289A, Asp291A | Helps bind and stabilise the intermediates. | electrostatic stabiliser |
Chemical Components
bimolecular nucleophilic substitution, overall reactant used, dephosphorylation, overall product formed, intermediate formation, proton transfer, rate-determining step, hydrolysis, intermediate collapse, intermediate terminated, native state of enzyme regeneratedReferences
- Murray B et al. (2016), Proc Natl Acad Sci U S A, 113, 2104-2109. Crystallography captures catalytic steps in human methionine adenosyltransferase enzymes. DOI:10.1073/pnas.1510959113. PMID:26858410.
- Liu Y et al. (2016), Frontiers of Chemical Science and Engineering, 10, 238-244. Functional characterization of a thermostable methionine adenosyltransferase from Thermus thermophilus HB27. DOI:10.1007/s11705-016-1566-2.
- Markham GD et al. (2009), Arch Biochem Biophys, 492, 82-92. An investigation of the catalytic mechanism of S-adenosylmethionine synthetase by QM/MM calculations. DOI:10.1016/j.abb.2009.08.010. PMID:19699176.
- Komoto J et al. (2004), Biochemistry, 43, 1821-1831. Crystal Structure of theS-Adenosylmethionine Synthetase Ternary Complex: A Novel Catalytic Mechanism ofS-Adenosylmethionine Synthesis from ATP and Met†,‡. DOI:10.1021/bi035611t. PMID:14967023.
- Taylor JC et al. (1999), J Biol Chem, 274, 32909-32914. The Bifunctional Active Site of S-Adenosylmethionine Synthetase: ROLES OF THE ACTIVE SITE ASPARTATES. DOI:10.1074/jbc.274.46.32909. PMID:10551856.
- Takusagawa F et al. (1996), Biochemistry, 35, 2586-2596. Structure and Function ofS-Adenosylmethionine Synthetase: Crystal Structures ofS-Adenosylmethionine Synthetase with ADP, BrADP, and PPiat 2.8 Å Resolution†,‡. DOI:10.1021/bi952604z. PMID:8611562.
- Takusagawa F et al. (1996), J Biol Chem, 271, 136-147. Crystal structure of S-adenosylmethionine synthetase. DOI:10.2210/pdb1xra/pdb. PMID:8550549.
- Fu Z et al. (1996), J Biomol Struct Dyn, 13, 727-739. Flexible Loop in the Structure of S-Adenosylmethionine Synthetase Crystallized in the Tetragonal Modification. DOI:10.1080/07391102.1996.10508887. PMID:8723769.
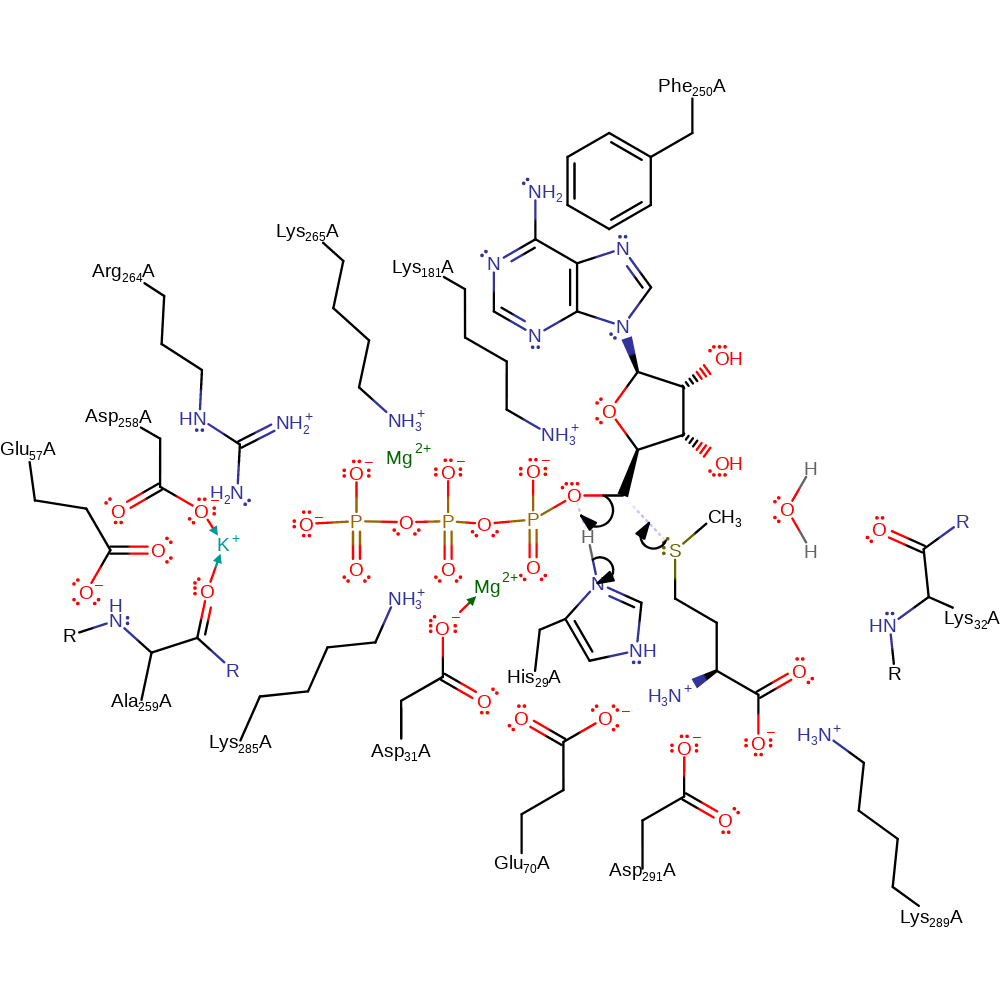
Step 1. The C5'-O5' bond of ATP dissociates, the phosphate of the leaving group deprotonates His29, the anionic histidine is stabilised by the main chain amides of Lys32 and Asp31. A simultaneous change in the ribose ring conformation from C4'-exo to C3'-endo occurs, and the SD of methionine makes a nucleophilic attack on the C5' to form S-adenosylmethionine. His29 deprotonates the leaving group in an overall nucleophilic substitution reaction.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu57A | metal ligand |
| Ala259A (main-C) | metal ligand |
| Asp258A | metal ligand, electrostatic stabiliser, steric role |
| Glu70A | electrostatic stabiliser, steric role |
| Lys289A | electrostatic stabiliser |
| Phe250A | steric role |
| Lys285A | electrostatic stabiliser |
| Lys265A | electrostatic stabiliser |
| Lys181A | electrostatic stabiliser |
| Arg264A | electrostatic stabiliser |
| Asp31A | metal ligand, electrostatic stabiliser |
| Lys32A (main-N) | electrostatic stabiliser |
| Asp291A | electrostatic stabiliser |
| His29A | proton donor |
Chemical Components
ingold: bimolecular nucleophilic substitution, overall reactant used, dephosphorylation, overall product formed, intermediate formation, proton transfer, rate-determining stepCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp31A | electrostatic stabiliser |
| Lys32A (main-N) | electrostatic stabiliser |
| Lys181A | electrostatic stabiliser |
| Lys289A | electrostatic stabiliser |
| Lys285A | electrostatic stabiliser |
| Lys265A | electrostatic stabiliser |
| Arg264A | electrostatic stabiliser |
| Glu57A | metal ligand |
| Ala259A (main-C) | metal ligand |
| Asp258A | metal ligand |
| Asp31A | metal ligand |
| Asp291A | electrostatic stabiliser |
| His29A | proton acceptor |






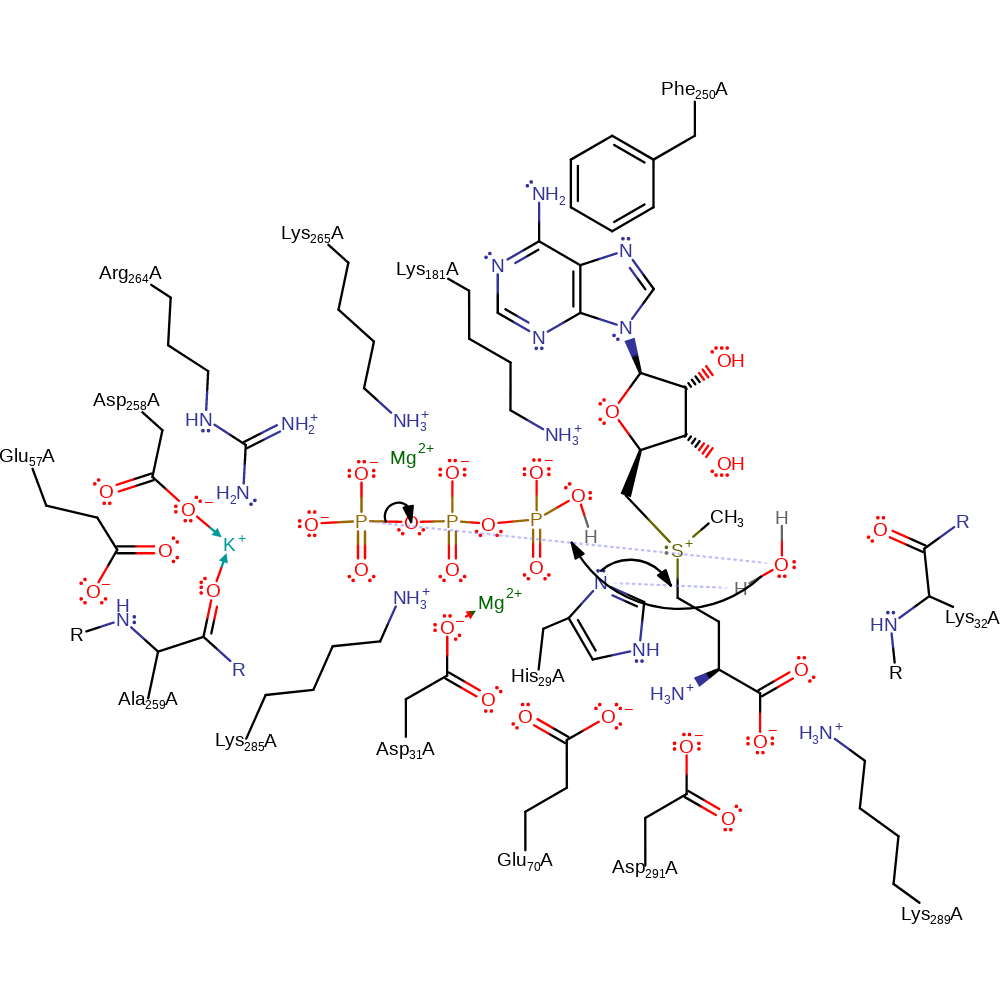
 Download:
Download: