Phosphogluconate dehydrogenase (decarboxylating)
The enzyme participates in the oxidative branch of the pentose phosphate pathway, whose main purpose is to produce NADPH and pentose for biosynthetic reactions. It catalyses the oxidative decarboxylation of 6-phosphogluconate to ribulose 5-phosphate and CO2, with concomitant reduction of NADP to NADPH.
Reference Protein and Structure
- Sequence
-
P00349
 (1.1.1.44)
(1.1.1.44)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Ovis aries (sheep)

- PDB
-
2pgd
- THE STRUCTURE OF 6-PHOSPHOGLUCONATE DEHYDROGENASE REFINED AT 2 ANGSTROMS RESOLUTION
(2.0 Å)



- Catalytic CATH Domains
-
1.10.1040.10
 3.40.50.720
3.40.50.720  (see all for 2pgd)
(see all for 2pgd)
Enzyme Reaction (EC:1.1.1.44)
Enzyme Mechanism
Introduction
The general base accepts a proton from the 3-hydroxyl group of 6PG concomitant with hydride transfer and then shuttles the proton between itself and the sugar oxygen throughout the reaction, ultimately accepting it as ribulose is formed. The general acid presumably plays a role in only the last of the three steps, namely, the tautomerisation of the enediol of ribulose 5-phosphate to the keto product.
Catalytic Residues Roles
| UniProt | PDB* (2pgd) | ||
| His187, Ser129, Asn188 | His186A, Ser128A, Asn187A | This triad(1) performs three different roles during the course of the reaction. It participates in the precatalytic conformational change; provides ground state binding affinity for 6PG and NADPH; and finally, affects the relative rates of reduction or decarboxylation of the 3-keto-6PG intermediate. This rate enhancement is achieved by anchoring the cofactor after hydride transfer, accompanied by the rotation of the nicotinamide ring around the N-glycosidic bond and displacement of C1 of 6PG. | promote heterolysis, electrostatic stabiliser |
| Lys184, Glu191 | Lys183A, Glu190A | Acts as a general acid/base. | proton acceptor, proton donor |
Chemical Components
proton transfer, hydride transfer, overall reactant used, decarboxylation, unimolecular elimination by the conjugate base, overall product formed, assisted keto-enol tautomerisation, inferred reaction step, native state of enzyme regeneratedReferences
- Li L et al. (2006), Biochemistry, 45, 12680-12686. Role of the S128, H186, and N187 triad in substrate binding and decarboxylation in the sheep liver 6-phosphogluconate dehydrogenase reaction. DOI:10.1021/bi0613675. PMID:17042485.
- Hanau S et al. (2014), Biochim Biophys Acta, 1844, 785-792. Energy cost for the proper ionization of active site residues in 6-phosphogluconate dehydrogenase from T. brucei. DOI:10.1016/j.bbapap.2014.02.011. PMID:24568863.
- Chen YY et al. (2010), J Struct Biol, 169, 25-35. Conformational changes associated with cofactor/substrate binding of 6-phosphogluconate dehydrogenase from Escherichia coli and Klebsiella pneumoniae: Implications for enzyme mechanism. DOI:10.1016/j.jsb.2009.08.006. PMID:19686854.
- Cervellati C et al. (2008), Biochemistry, 47, 1862-1870. Proper orientation of the nicotinamide ring of NADP is important for the precatalytic conformational change in the 6-phosphogluconate dehydrogenase reaction. DOI:10.1021/bi7015684. PMID:18205398.
- He W et al. (2007), BMC Struct Biol, 7, 38-. Crystal structure of Saccharomyces cerevisiae 6-phosphogluconate dehydrogenase Gnd1. DOI:10.1186/1472-6807-7-38. PMID:17570834.
- Sundaramoorthy R et al. (2007), FEBS J, 274, 275-286. Crystal structures of a bacterial 6-phosphogluconate dehydrogenase reveal aspects of specificity, mechanism and mode of inhibition by analogues of high-energy reaction intermediates. DOI:10.1111/j.1742-4658.2006.05585.x. PMID:17222187.
- Wang J et al. (2006), J Phys Chem B, 110, 7029-7035. Catalytic mechanism of 6-phosphogluconate dehydrogenase: a theoretical investigation. DOI:10.1021/jp0564748. PMID:16571018.
- Cervellati C et al. (2005), Biochemistry, 44, 2432-2440. Role of methionine-13 in the catalytic mechanism of 6-phosphogluconate dehydrogenase from sheep liver. DOI:10.1021/bi0476679. PMID:15709755.
- Dardonville C et al. (2004), J Med Chem, 47, 3427-3437. Selective inhibition of Trypanosoma brucei 6-phosphogluconate dehydrogenase by high-energy intermediate and transition-state analogues. DOI:10.1021/jm031066i. PMID:15189039.
- Hanau S et al. (2004), Curr Med Chem, 11, 2639-2650. 6-phosphogluconate dehydrogenase: a target for drugs in African trypanosomes. PMID:15544466.
- Zhang L et al. (1999), Biochemistry, 38, 11231-11238. Lysine 183 is the general base in the 6-phosphogluconate dehydrogenase-catalyzed reaction. DOI:10.1021/bi990433i. PMID:10471272.
- Rippa M et al. (1998), Biochim Biophys Acta, 1429, 83-92. 6-Phosphogluconate dehydrogenase: the mechanism of action investigated by a comparison of the enzyme from different species. PMID:9920387.
- Karsten WE et al. (1998), Biochemistry, 37, 15691-15697. Glutamate 190 is a general acid catalyst in the 6-phosphogluconate-dehydrogenase-catalyzed reaction. DOI:10.1021/bi9812827. PMID:9843373.
- Phillips C et al. (1998), J Mol Biol, 282, 667-681. A 2.8 A resolution structure of 6-phosphogluconate dehydrogenase from the protozoan parasite Trypanosoma brucei: comparison with the sheep enzyme accounts for differences in activity with coenzyme and substrate analogues. DOI:10.1006/jmbi.1998.2059. PMID:9737929.
- Hawes JW et al. (1996), FEBS Lett, 389, 263-267. Structural and mechanistic similarities of 6-phosphogluconate and 3-hydroxyisobutyrate dehydrogenases reveal a new enzyme family, the 3-hydroxyacid dehydrogenases. PMID:8766712.
- Price NE et al. (1996), Arch Biochem Biophys, 336, 215-223. Kinetic and chemical mechanisms of the sheep liver 6-phosphogluconate dehydrogenase. DOI:10.1006/abbi.1996.0551. PMID:8954568.
- Adams MJ et al. (1994), Structure, 2, 651-668. Crystallographic study of coenzyme, coenzyme analogue and substrate binding in 6-phosphogluconate dehydrogenase: implications for NADP specificity and the enzyme mechanism. PMID:7922042.
- Berdis AJ et al. (1993), Biochemistry, 32, 2041-2046. Chemical mechanism of 6-phosphogluconate dehydrogenase from Candida utilis from pH studies. DOI:10.1021/bi00059a022.
- Adams MJ et al. (1991), Acta Crystallogr B, 47 ( Pt 5), 817-820. The structure of 6-phosphogluconate dehydrogenase refined at 2.5 A resolution. PMID:1793548.
- Adams MJ et al. (1983), EMBO J, 2, 1009-1014. The three dimensional structure of sheep liver 6-phosphogluconate dehydrogenase at 2.6 A resolution. PMID:6641716.
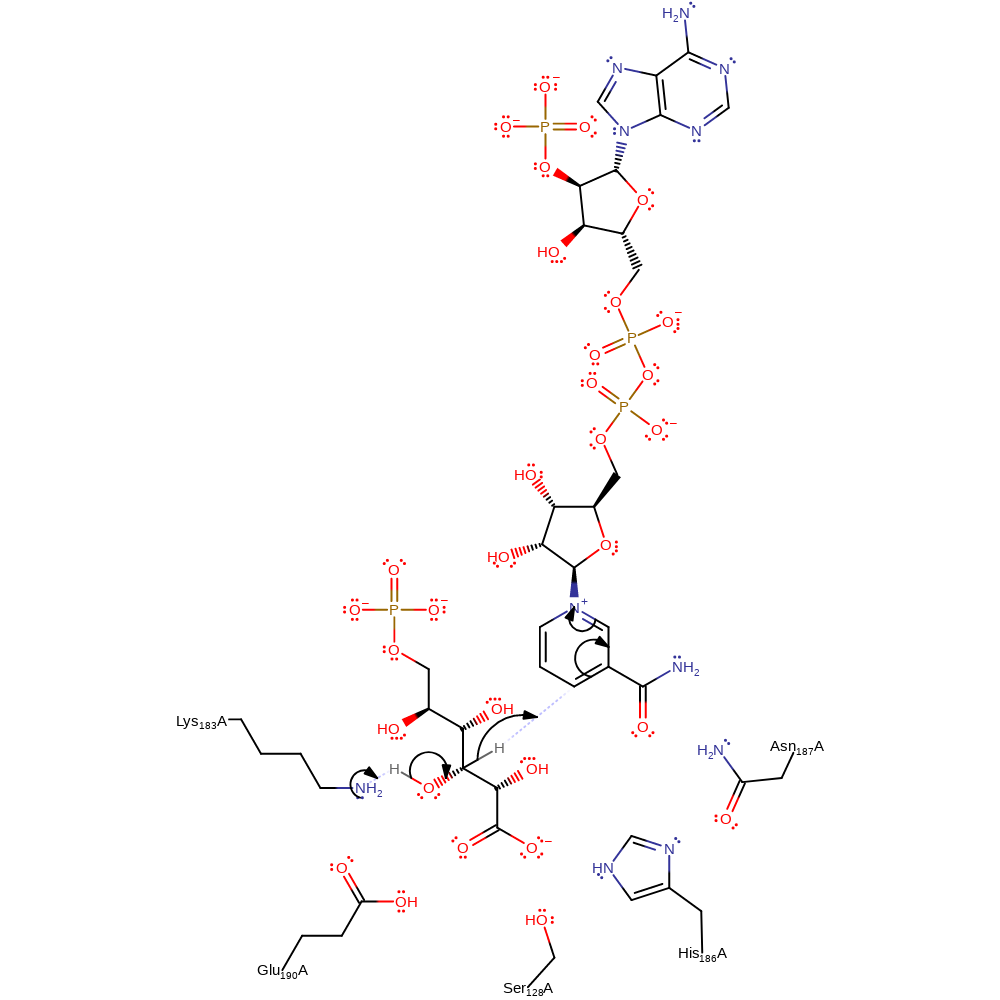
Step 1. Lys183 abstracts a proton from the sugar substrate, eliminating a hydride ion, which is added to the NADP cosubstrate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Ser128A | electrostatic stabiliser |
| His186A | electrostatic stabiliser |
| Asn187A | electrostatic stabiliser |
| Lys183A | proton acceptor |
Chemical Components
proton transfer, hydride transfer, overall reactant used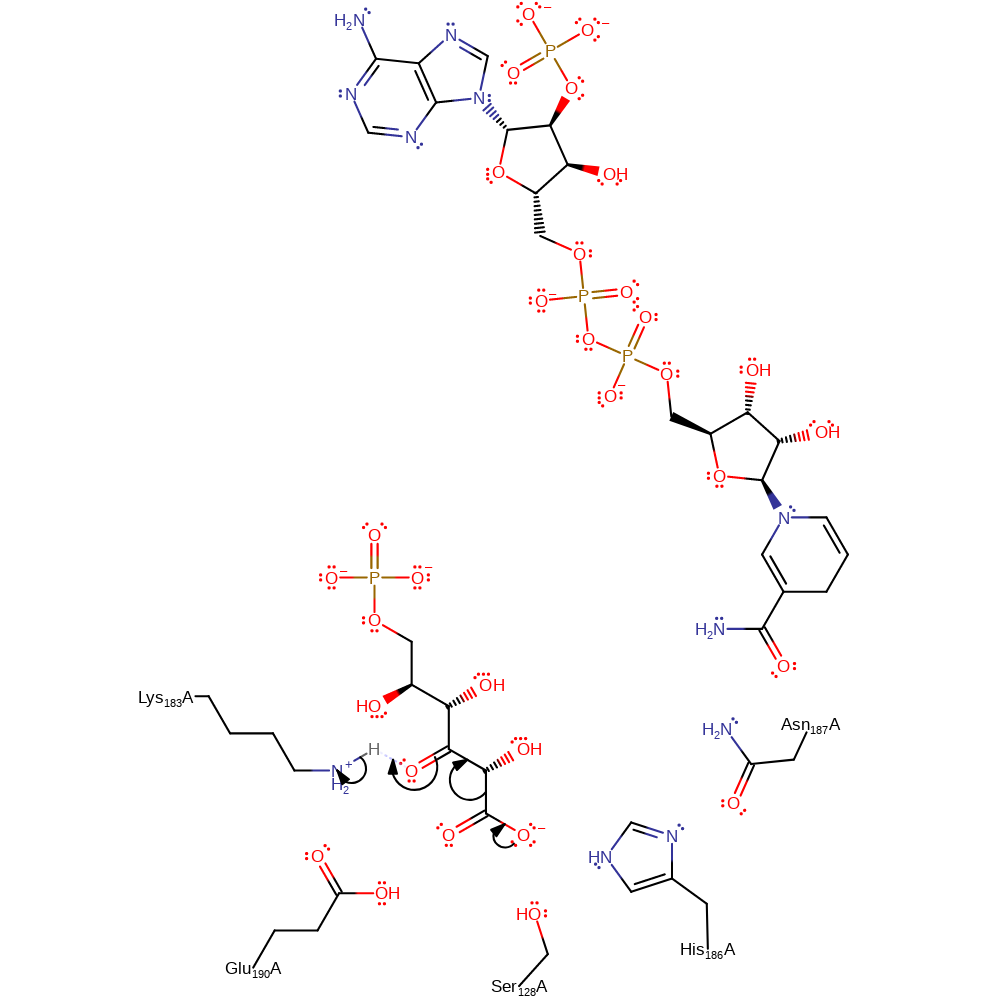
Step 2. The NADPH rotates away from the active centre. The substrate than undergoes decarboxylation and Lys183 donates a proton back to the intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Ser128A | promote heterolysis |
| His186A | promote heterolysis |
| Asn187A | promote heterolysis |
| Lys183A | proton donor |
Chemical Components
decarboxylation, ingold: unimolecular elimination by the conjugate base, proton transfer, overall product formed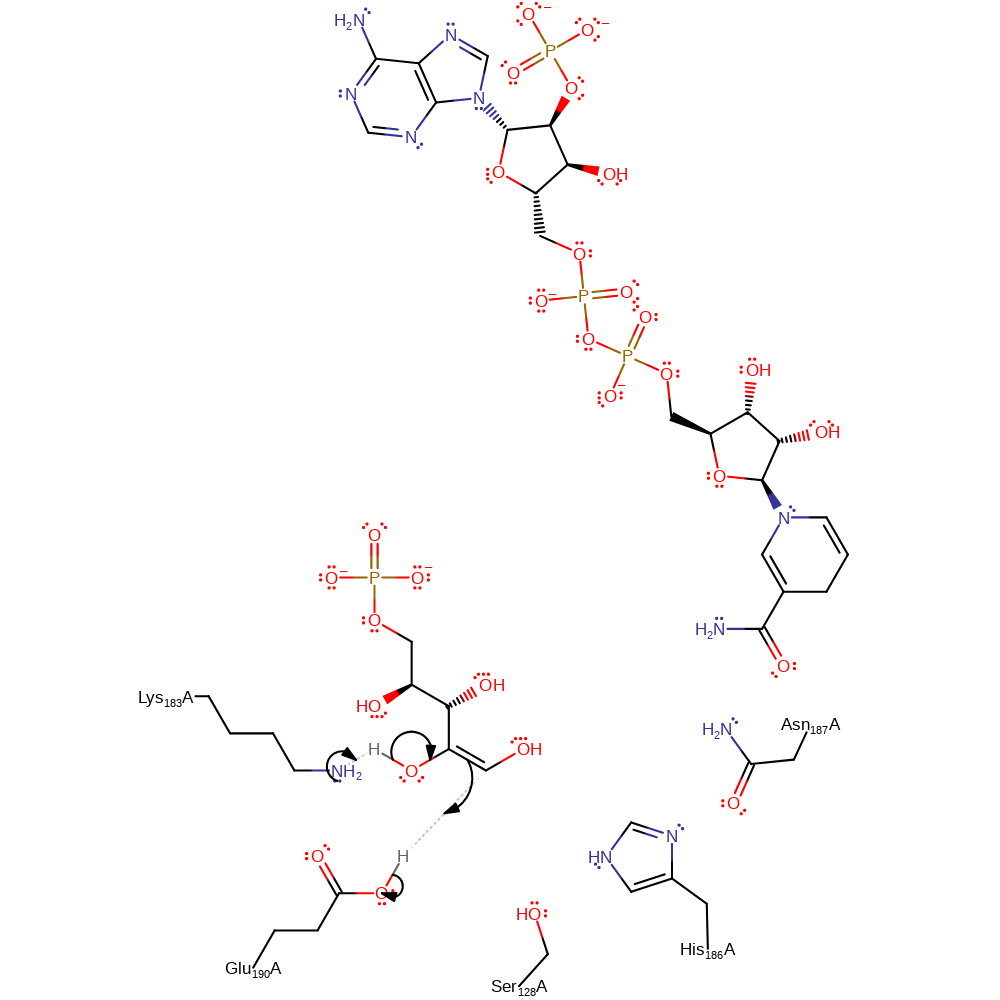
Step 3. Carbon dioxide diffuses from the active site. Lys183 initiates an assisted keto-enol tautomerisation to form the final product which diffuses from the active site along with NADPH.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu190A | proton donor |
| Lys183A | proton acceptor |
Chemical Components
proton transfer, assisted keto-enol tautomerisation, overall product formed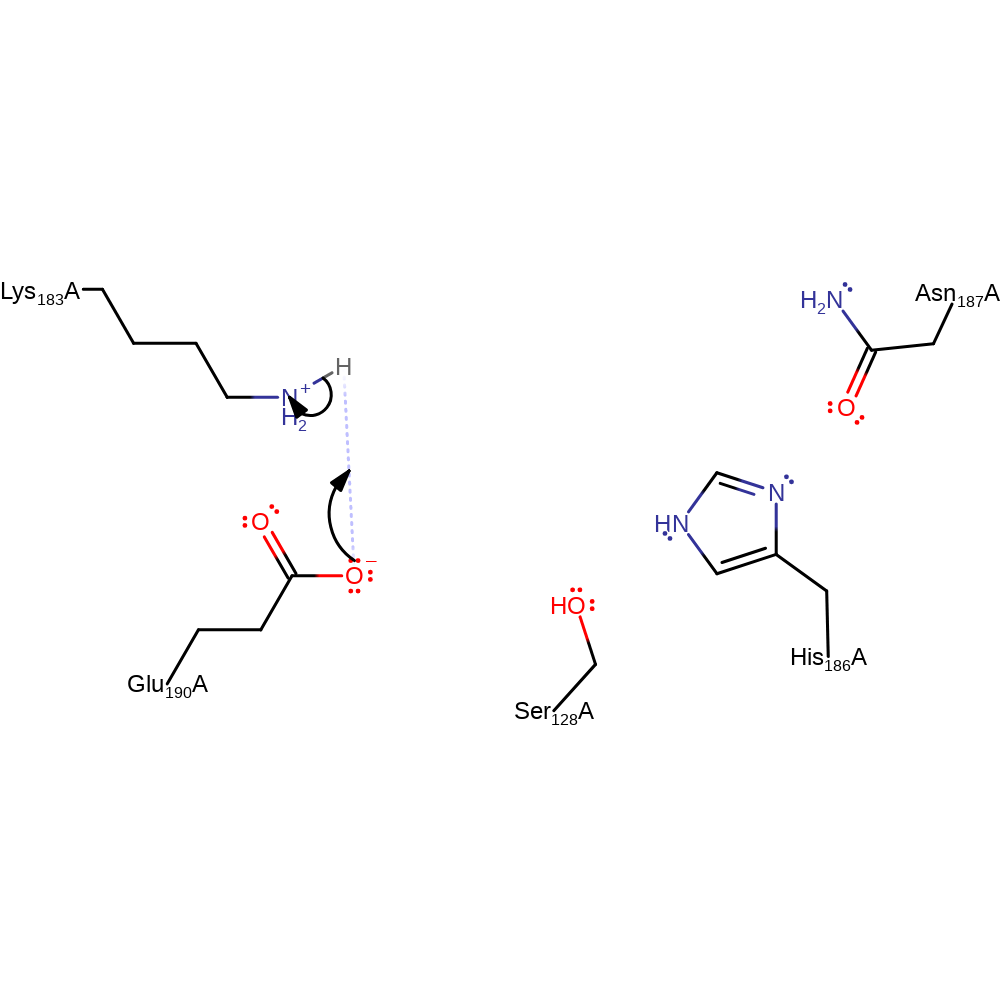
Step 4. Inferred return step to regenerate the active site protonation states. We assume that the proton can be directly transferred from Glu190 to Lys183.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys183A | proton donor |
| Glu190A | proton acceptor |





 Download:
Download: