Fructan beta-fructosidase
Exo-inulase from Aspergillus awamori (a fungus) is a member of the glycoside hydrolase family 32. It catalyses the hydrolysis of terminal, non-reducing 2,1- and 2,6-linked beta-D-fructofuranose residues in fructans. Fructans are used as food storage for some plant species, and exo-inulase is involved in their breakdown into fructose.
Reference Protein and Structure
- Sequence
-
Q96TU3
 (3.2.1.80)
(3.2.1.80)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Aspergillus awamori (Fungus)

- PDB
-
1y9m
- Crystal structure of exo-inulinase from Aspergillus awamori in spacegroup P212121
(1.89 Å)



- Catalytic CATH Domains
-
2.115.10.20
 (see all for 1y9m)
(see all for 1y9m)
Enzyme Reaction (EC:3.2.1.80)
Enzyme Mechanism
Introduction
Exo-inulase catalyses a double displacement mechanism, resulting in retention of configuration. Asp 41 acts as a nucleophile, attacking the carbon atom of the glycosidic bond. As the glycosidic bond is broken, Glu 241 acts as a general acid by donating a proton to the leaving group oxygen atom. Glu 241 then acts as a general base, deprotonating a water molecule, and activating it for nucleophilic attack on the carbon atom. As HO- nucleophilically attacks, Asp 41 is the leaving group.
Catalytic Residues Roles
| UniProt | PDB* (1y9m) | ||
| Asp41 | Asp41(22)A | Asp acts as a nucleophile, and attacks the carbon atom of the glycosidic bond. | covalently attached, nucleofuge, nucleophile |
| Glu241 | Glu241(222)A | Glu 241 donates a proton to the leaving group oxygen atom. It also deprotonates a water molecule, activating it for nucleophilic attack. | proton acceptor, proton donor, activator, increase nucleophilicity, promote heterolysis |
Chemical Components
overall product formed, overall reactant used, proton transfer, bimolecular nucleophilic substitution, intermediate formation, intermediate terminated, hydrolysisReferences
- Nagem RA et al. (2004), J Mol Biol, 344, 471-480. Crystal Structure of Exo-inulinase from Aspergillus awamori: The Enzyme Fold and Structural Determinants of Substrate Recognition. DOI:10.1016/j.jmb.2004.09.024. PMID:15522299.
- Holyavka M et al. (2016), Biocatal Biotransformation, 34, 1-17. Structural and functional properties of inulinases: A review. DOI:10.1080/10242422.2016.1196486.
- Pouyez J et al. (2012), Biochimie, 94, 2423-2430. First crystal structure of an endo-inulinase, INU2, from Aspergillus ficuum: discovery of an extra-pocket in the catalytic domain responsible for its endo-activity. DOI:10.1016/j.biochi.2012.06.020. PMID:22750808.
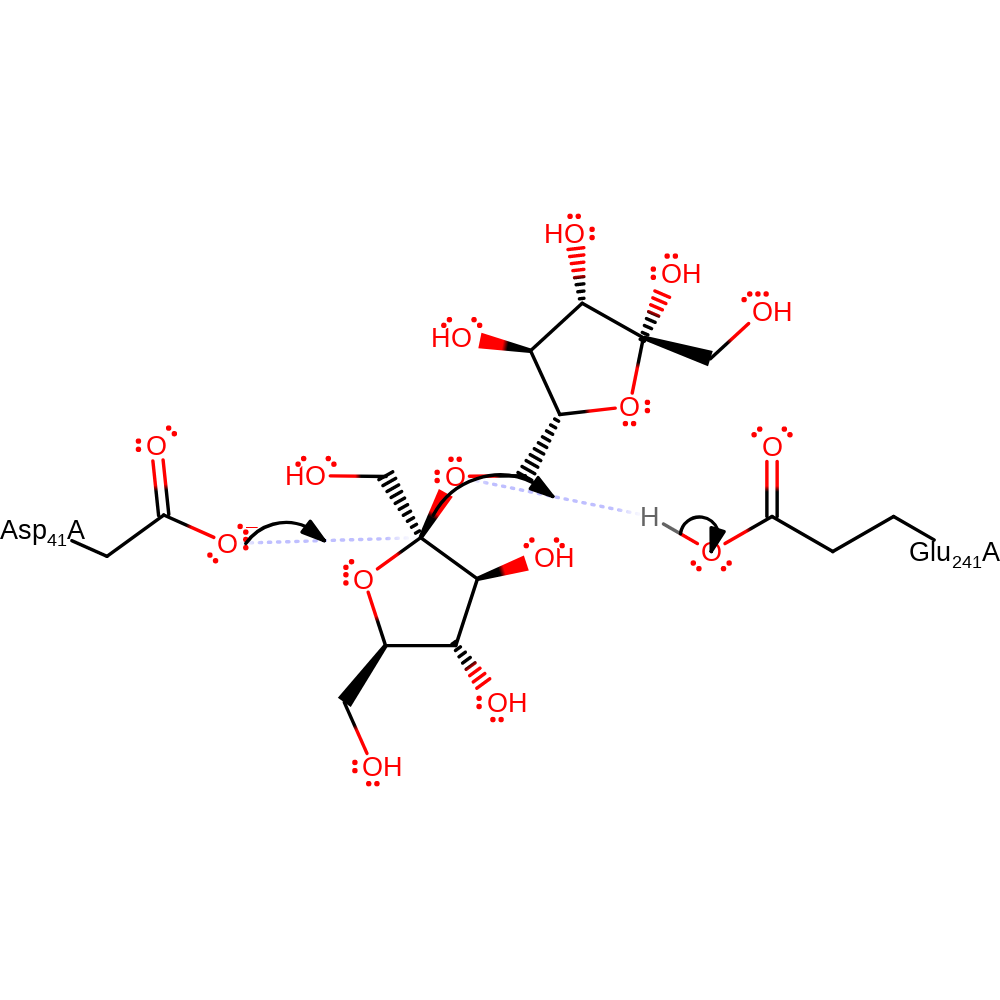
Step 1. Asp41 performs a nucleophilic attack on the glycosidic bond, while Glu241 protonates the leaving group. This leads to the formation of an enzyme-substrate intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp41(22)A | covalently attached |
| Glu241(222)A | promote heterolysis, proton donor |
| Asp41(22)A | nucleophile |
Chemical Components
overall product formed, overall reactant used, proton transfer, ingold: bimolecular nucleophilic substitution, intermediate formation
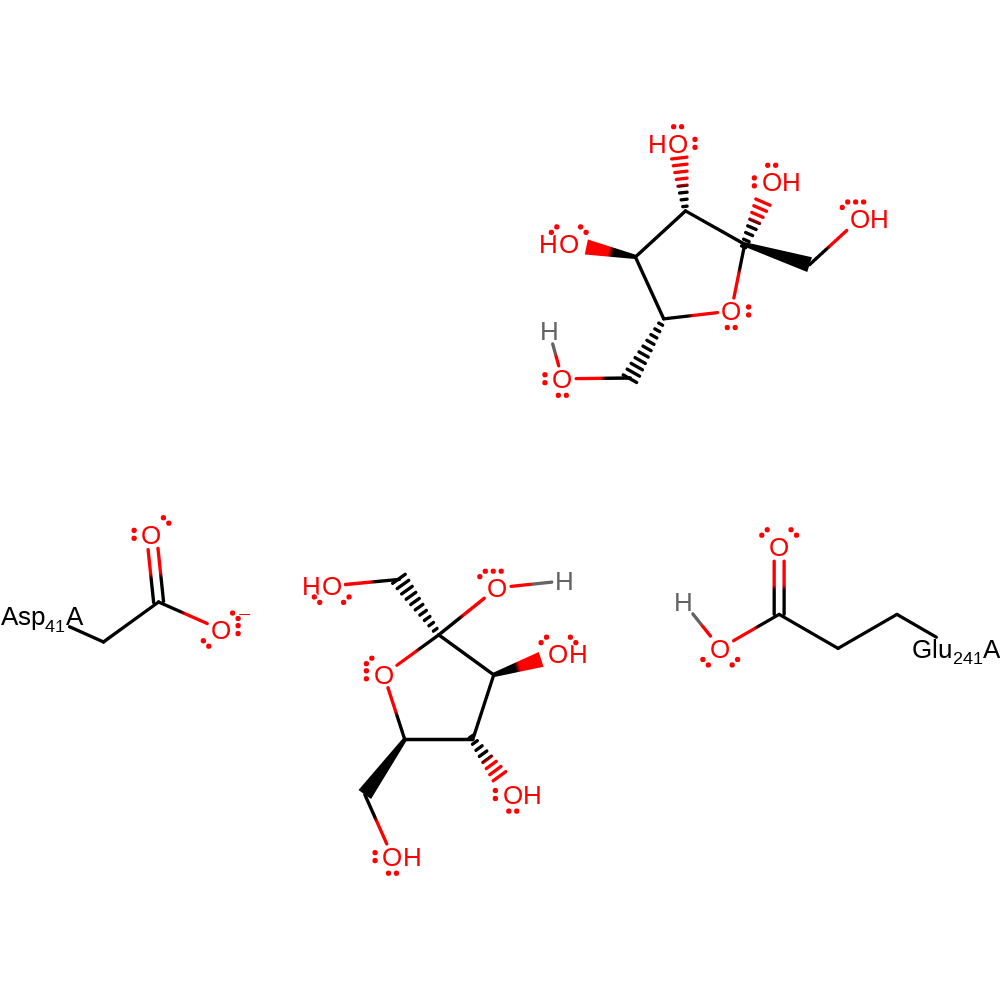
Step 2. Glu241 activates a water molecule which hydrolyses the intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu241(222)A | activator, increase nucleophilicity, proton acceptor |
| Asp41(22)A | nucleofuge |



 Download:
Download: