Mono-ADP-ribosyltransferase C3
Clostridium boltulinum C3 exoenzyme catalyses the transfer of the ADP-ribose moiety of NAD onto asparagine 41 of the small GTP-binding protein Rho. This ADP-ribosylation inactivates Rho, blocking Rho activity in regulating the cytoskeleton and leading to cytoskeleton depolymerisation. The protein synthesis inhibitors (inactivate the eukaryotic elongation factor 2 eEF2) diptheria toxin from Corynebacterium diphtheriae and exotoxin A (ETA) from Pseudomonas aeruginosa are also ADP-ribosyltransferases, contain a conserved Glu and thought to proceed through the same catalytic mechanism.
Reference Protein and Structure
- Sequence
-
P15879
 (2.4.2.-)
(2.4.2.-)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Clostridium botulinum D phage (Virus)

- PDB
-
1g24
- THE CRYSTAL STRUCTURE OF EXOENZYME C3 FROM CLOSTRIDIUM BOTULINUM
(1.7 Å)



- Catalytic CATH Domains
-
3.90.176.10
 (see all for 1g24)
(see all for 1g24)
Enzyme Mechanism
Introduction
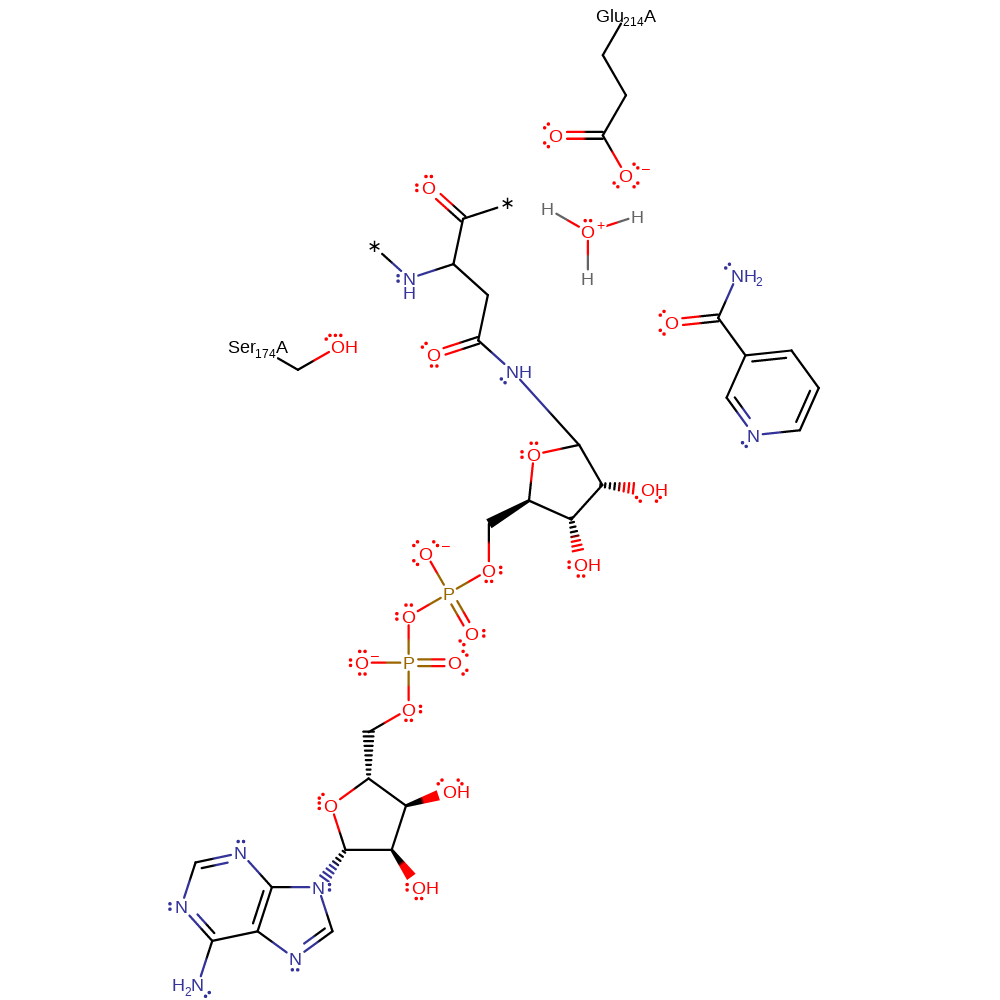
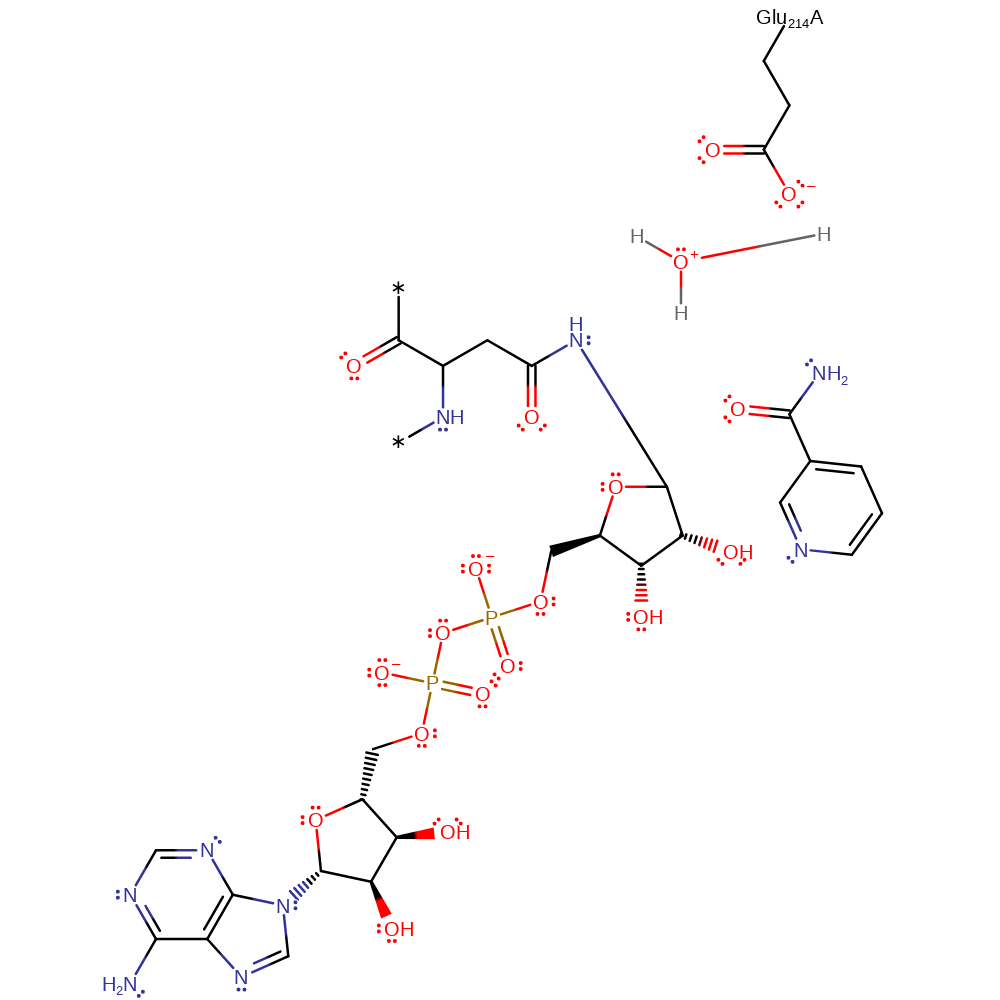
The reaction requires loss of the nicotinamide moiety of NAD and attack on the C1' of ribose by Asn 41 (thought to be activated by Glu214) of Rho in an Sn1 like mechanism. The oxocarbenium ion intermediate is thought to be stabilised by Glu 214, which forms a hydrogen bond to the ribose 2' OH group and also Ser174. Out of the two mechanism proposals, this is more favoured due to mutagenesis experiments of Glu214 uncoupling the glycohydrolytic and transferase reactions. It is thought Exotoxin A and the Diphtheria toxin share the same catalytic mechanism as the C3 exoenzyme.
Catalytic Residues Roles
| UniProt | PDB* (1g24) | ||
| Ser174 | Ser174(134)A | Stabilises the positive charge in the oxocarbenium-like transition state. Apart of the conserved STS motif. | electrostatic stabiliser, polar interaction |
| Glu214 | Glu214(174)A | Stabilises positive charge in the oxocarbenium-like intermediate by forming a hydrogen bond to the 2' OH group of the ribose moiety. | proton acceptor, proton donor, electrostatic stabiliser, polar interaction |
Chemical Components
heterolysis, intermediate formation, elimination (not covered by the Ingold mechanisms), overall product formed, bimolecular nucleophilic addition, overall reactant used, proton transfer, intermediate terminated, inferred reaction step, native state of enzyme regeneratedReferences
- Holbourn KP et al. (2006), FEBS J, 273, 4579-4593. A family of killer toxins. Exploring the mechanism of ADP-ribosylating toxins. DOI:10.1111/j.1742-4658.2006.05442.x. PMID:16956368.
- Vogelsgesang M et al. (2006), Biochemistry, 45, 1017-1025. Exchange of glutamine-217 to glutamate of Clostridium limosum exoenzyme C3 turns the asparagine-specific ADP-ribosyltransferase into an arginine-modifying enzyme. DOI:10.1021/bi052253g. PMID:16411778.
- Jørgensen R et al. (2005), Nature, 436, 979-984. Exotoxin A–eEF2 complex structure indicates ADP ribosylation by ribosome mimicry. DOI:10.1038/nature03871. PMID:16107839.
- Yates SP et al. (2004), Biochem J, 379, 563-572. Elucidation of eukaryotic elongation factor-2 contact sites within the catalytic domain of Pseudomonas aeruginosa exotoxin A. DOI:10.1042/bj20031731. PMID:14733615.
- Han S et al. (2001), J Mol Biol, 305, 95-107. Crystal structure and novel recognition motif of Rho ADP-ribosylating C3 exoenzyme from Clostridium botulinum: structural insights for recognition specificity and catalysis. DOI:10.1006/jmbi.2000.4292. PMID:11114250.
- Bell CE et al. (1997), Biochemistry, 36, 481-488. Crystal Structure of Nucleotide-Free Diphtheria Toxin†,‡. DOI:10.1021/bi962214s. PMID:9012663.
- Wilson BA et al. (1990), Biochemistry, 29, 8643-8651. Active-site mutations of diphtheria toxin: effects of replacing glutamic acid-148 with aspartic acid, glutamine, or serine. DOI:10.1021/bi00489a021. PMID:1980208.
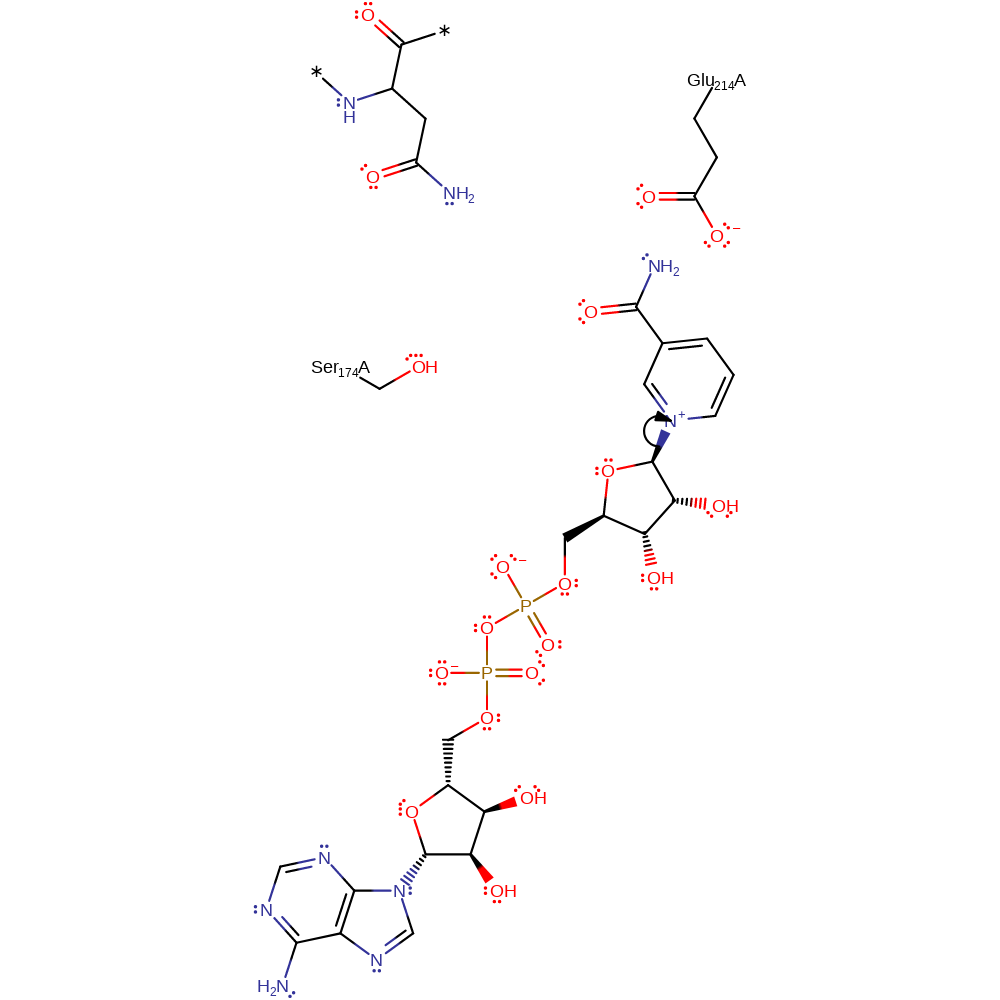
Step 1. Glycosidic bond cleavage resulting in an oxocarbenium intermediate, stabilised by Glu214 and Ser174.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu214(174)A | electrostatic stabiliser |
| Ser174(134)A | electrostatic stabiliser, polar interaction |
| Glu214(174)A | polar interaction |
Chemical Components
heterolysis, intermediate formation, elimination (not covered by the Ingold mechanisms), overall product formed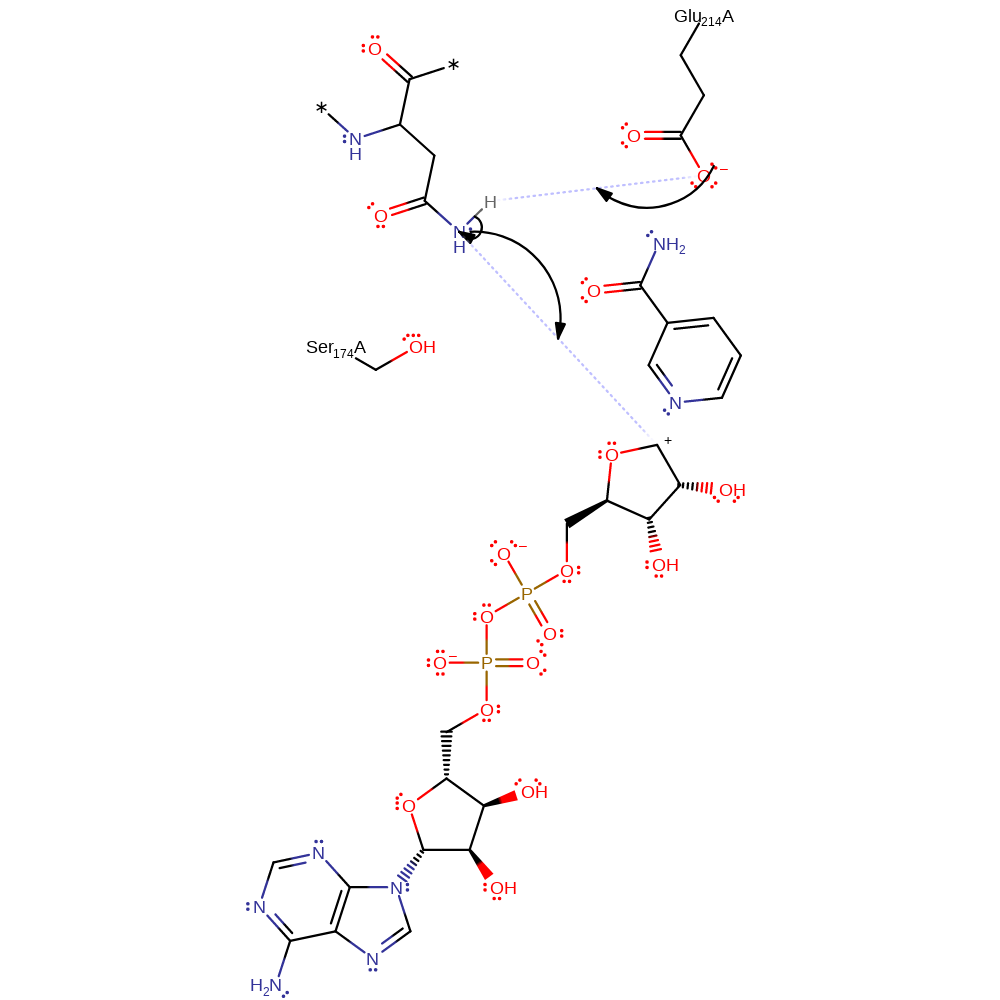
Step 2. Nucleophilic attack by Asn41 on the Rho protein. Glu214 is proposed to deprotonate the arginine to increase it's nucleophilicity.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Ser174(134)A | electrostatic stabiliser |
| Glu214(174)A | electrostatic stabiliser |
| Ser174(134)A | polar interaction |
| Glu214(174)A | polar interaction, proton acceptor |
Chemical Components
ingold: bimolecular nucleophilic addition, overall reactant used, proton transfer, overall product formed, intermediate terminatedCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu214(174)A | proton donor |
Chemical Components
inferred reaction step, proton transfer, native state of enzyme regeneratedIntroduction
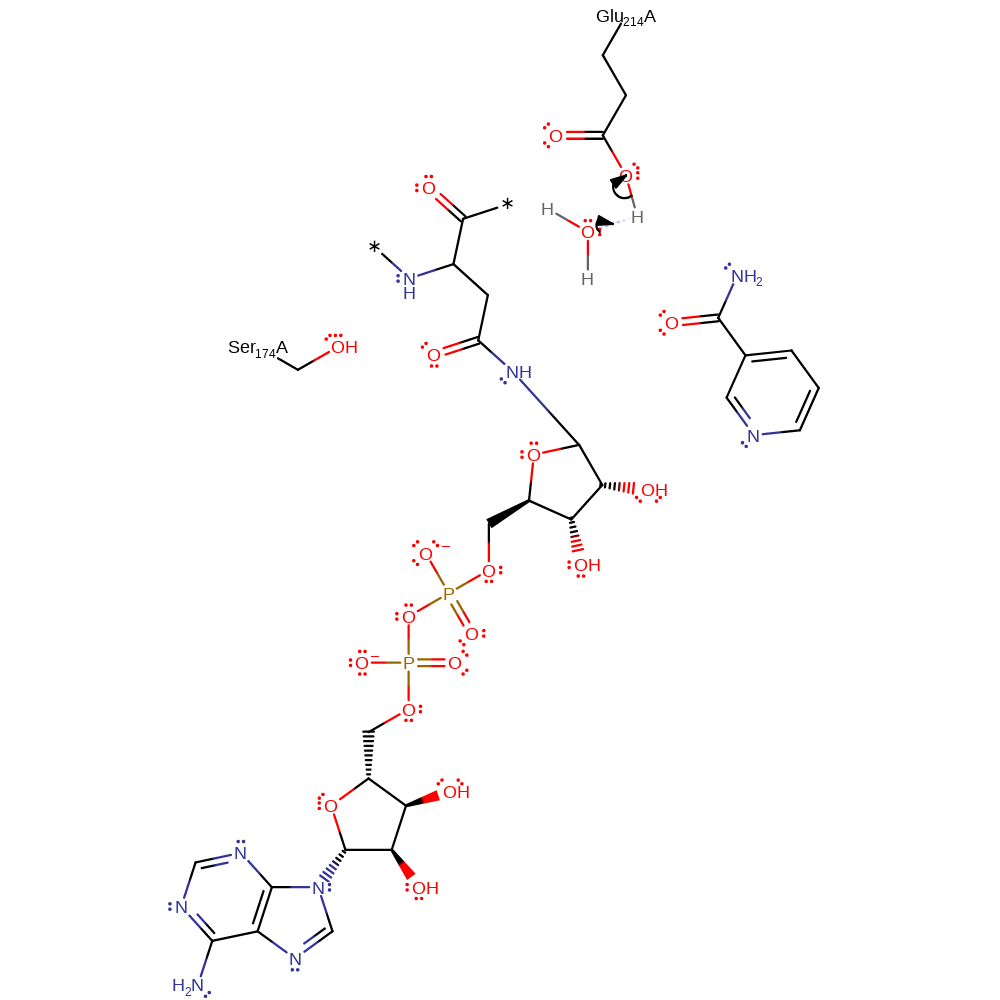
In an Sn2 manner, Asn41 is deprotonated and acts as a nucleophile, attacking C1 of the ribose sugar to generate an oxocarbenium ion like transition state. Glu214 hydrogen bonding to the nicotinamide ribose 2' OH increases the ring's electronegativity, stabilising the oxocarbenium ion thus facilitating glycosidic bond cleavage. The phosphates of NAD itself potentially increase the electrophilicity of C1 due to withdrawing electrons from the nicotinamide ring.
Catalytic Residues Roles
| UniProt | PDB* (1g24) | ||
| Glu214 | Glu214(174)A | Stabilises positive charge in the oxocarbenium-like transition state by forming a hydrogen bond to the 2' OH group of the ribose moiety. | proton acceptor, proton donor, electrostatic stabiliser, polar interaction |
Chemical Components
bimolecular nucleophilic substitution, overall reactant used, overall product formed, inferred reaction step, proton transfer, native state of enzyme regeneratedReferences
- Han S et al. (2001), J Mol Biol, 305, 95-107. Crystal structure and novel recognition motif of Rho ADP-ribosylating C3 exoenzyme from Clostridium botulinum: structural insights for recognition specificity and catalysis. DOI:10.1006/jmbi.2000.4292. PMID:11114250.
- Holbourn KP et al. (2006), FEBS J, 273, 4579-4593. A family of killer toxins. Exploring the mechanism of ADP-ribosylating toxins. DOI:10.1111/j.1742-4658.2006.05442.x. PMID:16956368.

Step 1. Glu214 is proposed to deprotonate Asn41 so that Asn41 nucleophilically attacks the C1 on the ribose ring.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu214(174)A | electrostatic stabiliser |
| Glu214(174)A | polar interaction, proton acceptor |
Chemical Components
ingold: bimolecular nucleophilic substitution, overall reactant used, overall product formedCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu214(174)A | polar interaction, proton donor |





 Download:
Download: 
 Download:
Download: