Omptin
Outer membrane protease (OmpT) of Escherichia coli is from a family of homologous outer membrane proteases called omptins. They are implicated in the virulence of several pathogenic, gram negative bacteria, and OmpT is associated with urinary tract disease. OmpT preferentially cleaves substrates between two consecutive basic amino acids, and is dependent on the presence of lipopolysaccharide.
Reference Protein and Structure
- Sequence
-
P09169
 (3.4.23.49)
(3.4.23.49)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Escherichia coli K-12 (Bacteria)

- PDB
-
1i78
- CRYSTAL STRUCTURE OF OUTER MEMBRANE PROTEASE OMPT FROM ESCHERICHIA COLI
(2.6 Å)



- Catalytic CATH Domains
-
2.40.128.90
 (see all for 1i78)
(see all for 1i78)
Enzyme Reaction (EC:3.4.23.49)
Enzyme Mechanism
Introduction
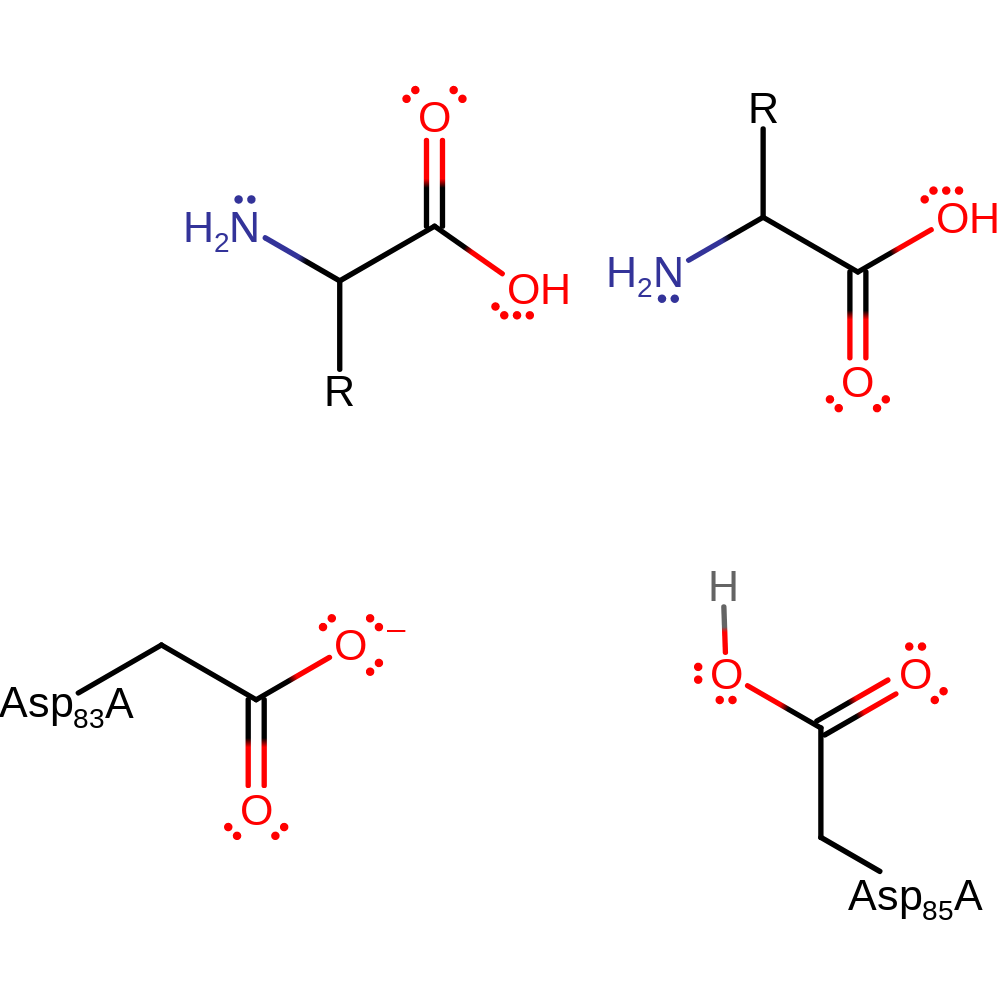
To initiate the reaction Asp 103 deprotonates a water which activates it to nucleophilically attack the carbonyl carbon. In addition Asp 105 protonates the carbonyl oxygen which results in the formation of the gemdiol intermediate. Asp 103 then protonates the amide nitrogen which weakens the scissile bond and thus when Asp 105 accepts a proton from the gemdiol hydroxyl this results in the collapse of the tetrahedral intermediate and the cleavage of the peptide bond.
Catalytic Residues Roles
| UniProt | PDB* (1i78) | ||
| Asp103 | Asp83A | Acts as a general acid/base, deprotonates water to activate it for nucleophilic attack and also protonates the amide nitrogen to weaken the scissile bond. | proton acceptor, proton donor |
| Asp105 | Asp85A | Acts as a general acid/base as protonates the carbonyl oxygen to form the gemdiol intermediate and also then deprotonates the hydroxyl to initiate an elimination and the cleavage of the peptide bond. | proton acceptor, proton donor |
Chemical Components
proton transfer, bimolecular nucleophilic addition, intermediate formation, overall reactant used, unimolecular elimination by the conjugate base, heterolysis, intermediate collapse, overall product formed, native state of enzyme regeneratedReferences
- Vandeputte-Rutten L et al. (2001), EMBO J, 20, 5033-5039. Crystal structure of the outer membrane protease OmpT from Escherichia coli suggests a novel catalytic site. DOI:10.1093/emboj/20.18.5033. PMID:11566868.
- Hritonenko V et al. (2007), Mol Membr Biol, 24, 395-406. Omptin proteins: an expanding family of outer membrane proteases in Gram-negative Enterobacteriaceae. DOI:10.1080/09687680701443822. PMID:17710644.
- Baaden M et al. (2004), Biophys J, 87, 2942-2953. OmpT: Molecular Dynamics Simulations of an Outer Membrane Enzyme. DOI:10.1529/biophysj.104.046987. PMID:15315948.
- Vandeputte-Rutten L (2002), Curr Opin Struct Biol, 12, 704-708. Novel proteases: common themes and surprising features. DOI:10.1016/S0959-440X(02)00393-7.

Step 1. Asp103 deprotonates a water which activates it to nucleophilically attack the carbon of the carbonyl. Simultaneously Asp 105 protonates the carbonyl oxygen. This results in a gemdiol intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp83A | proton acceptor |
| Asp85A | proton donor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, intermediate formation, overall reactant used
Step 2. Asp103 protonates the the amide nitrogen of the scissile peptide bond, which is simultaneously and progressively weakened and elongated.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp83A | proton donor |
Chemical Components
proton transfer, intermediate formation
Step 3. Asp105 deprotonates a hydroxyl of the gemdiol intermediate awhich initiates an elimination which results in the cleavage of the peptide bond.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp85A | proton acceptor |
Chemical Components
proton transfer, ingold: unimolecular elimination by the conjugate base, heterolysis, intermediate collapse, overall product formed, native state of enzyme regeneratedIntroduction
In this mechanism the water molecule acts in a similar way to the serine in a serine protease. The water is deprotonated by His 232 which is hydrogen bonded to Asp 230 and this will lower the pKa of His 232 so it more willingly accepts a proton from water. Once deprotonated the water will act as a nucleophile and attack the carbonyl carbon to produce an oxyanion intermediate and the negative charge will be stabilised by the oxyanion hole comprised of the amide backbones of Asp 103 and Asp 105. His 232 will protonate the amide nitrogen which causes the tetrahedral intermediate to collapse and this results in the cleavage of the peptide bond and the release of the products.
Catalytic Residues Roles
| UniProt | PDB* (1i78) | ||
| Asp230 | Asp210A | Hydrogen bonds to His 232 which stabilises Histidine and lowers its pKa so it more willingly accepts a proton from water. | electrostatic stabiliser |
| His232 | His212A | Acts as a general acid/base as accepts a proton from water to activate it for nucleophilic attack and then transfers the proton to the amide nitrogen to initiate an elimination. | proton acceptor, proton donor |
| Asp103 (main-N), Asp105 (main-N) | Asp83A (main-N), Asp85A (main-N) | Form the oxyanion hole and stabilise the negative charge on the oxygen of the tetrahedral intermediate through hydrogen bonding. | electrostatic stabiliser |
Chemical Components
proton transfer, bimolecular nucleophilic addition, intermediate formation, overall reactant used, unimolecular elimination by the conjugate base, heterolysis, intermediate collapse, overall product formed, native state of enzyme regeneratedReferences
- Vandeputte-Rutten L et al. (2001), EMBO J, 20, 5033-5039. Crystal structure of the outer membrane protease OmpT from Escherichia coli suggests a novel catalytic site. DOI:10.1093/emboj/20.18.5033. PMID:11566868.
- Hritonenko V et al. (2007), Mol Membr Biol, 24, 395-406. Omptin proteins: an expanding family of outer membrane proteases in Gram-negative Enterobacteriaceae. DOI:10.1080/09687680701443822. PMID:17710644.
- Baaden M et al. (2004), Biophys J, 87, 2942-2953. OmpT: Molecular Dynamics Simulations of an Outer Membrane Enzyme. DOI:10.1529/biophysj.104.046987. PMID:15315948.
- Vandeputte-Rutten L (2002), Curr Opin Struct Biol, 12, 704-708. Novel proteases: common themes and surprising features. DOI:10.1016/S0959-440X(02)00393-7.

Step 1. His232 deprotonates a water which activates it to nucleophilically attack the carbonyl carbon.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp83A (main-N) | electrostatic stabiliser |
| Asp85A (main-N) | electrostatic stabiliser |
| Asp210A | electrostatic stabiliser |
| His212A | proton acceptor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, intermediate formation, overall reactant used
Step 2. His232 protonates the amide nitrogen which initiates an elimination from the oxyanion that results in the cleavage of the peptide bond.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp83A (main-N) | electrostatic stabiliser |
| Asp85A (main-N) | electrostatic stabiliser |
| Asp210A | electrostatic stabiliser |
| His212A | proton donor |
Chemical Components
proton transfer, ingold: unimolecular elimination by the conjugate base, heterolysis, intermediate collapse, overall product formed, native state of enzyme regeneratedIntroduction
Theoretical Mechanism. His 232 activates Ser 119 by acting as a general base. The protonated form of His 232 is stabilised through hydrogen bonding to Asp 230. Ser 119 nucleophilically attacks the scissile peptide bond, forming a negatively charged intermediate. This intermediate is stabilised by the oxyanion hole formed by proton sharing between Asp 103, Asp 105 and possibly a water molecule. When the carbonyl of the substrate is reformed, the leaving group is protonated by His 232. His 232 deprotonates a water molecule which activates it for nucleophilic attack on the carbonyl of the substrate. Another negatively charged, tetrahedral intermediate is formed. On reforming the carbonyl, Ser 119 is the leaving group, and is protonated by His 232.
Catalytic Residues Roles
| UniProt | PDB* (1i78) | ||
| Asp230 | Asp210A | Is thought to either stabilise the protonated form of His 212, or in acting as a base to activate water for nucleophilic attack. | electrostatic stabiliser |
| His232 | His212A | Activates Ser 99 by acting as a general base. Activates a water molecule again by acting as a general base. Provides leaving group with proton by acting as a general acid. | proton acceptor, proton donor |
| Ser119 | Ala99A | Hydroxyl side group is deprotonated by His 212. This then goes on to nucleophilically attack the scissile peptide bond of the substrate. | nucleofuge, nucleophile, proton acceptor, proton donor |
| Asp103 (main-N) | Asp83A (main-N) | Forms the oxyanion hole and stabilises the negatively charged intermediate via proton sharing (perhaps through a water molecule) with Asp 85. | electrostatic stabiliser |
| Asp105 (main-N) | Asp85A (main-N) | Forms the oxyanion hole and stabilises the negatively charged intermediate via proton sharing (perhaps through a water molecule) with Asp 83. | electrostatic stabiliser |
Chemical Components
proton transfer, bimolecular nucleophilic addition, enzyme-substrate complex formation, intermediate formation, overall reactant used, unimolecular elimination by the conjugate base, heterolysis, intermediate collapse, overall product formed, enzyme-substrate complex cleavage, native state of enzyme regeneratedReferences
- Vandeputte-Rutten L et al. (2001), EMBO J, 20, 5033-5039. Crystal structure of the outer membrane protease OmpT from Escherichia coli suggests a novel catalytic site. DOI:10.1093/emboj/20.18.5033. PMID:11566868.
- Baaden M et al. (2004), Biophys J, 87, 2942-2953. OmpT: Molecular Dynamics Simulations of an Outer Membrane Enzyme. DOI:10.1529/biophysj.104.046987. PMID:15315948.
- Kramer RA et al. (2000), FEBS Lett, 468, 220-224. Identification of active site serine and histidine residues inEscherichia coliouter membrane protease OmpT. DOI:10.1016/s0014-5793(00)01231-x. PMID:10692590.

Step 1. His232 deprotonates Ser119 which activates it to nucleophilically attack the carbon of the carbonyl group.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp85A (main-N) | electrostatic stabiliser |
| Asp83A (main-N) | electrostatic stabiliser |
| Asp210A | electrostatic stabiliser |
| Ala99A | nucleophile |
| His212A | proton acceptor |
| Ala99A | proton donor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, enzyme-substrate complex formation, intermediate formation, overall reactant used
Step 2. His232 protonates the amide which initiates an elimination from the oxyanion which results in the cleavage of the peptide bond.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp83A (main-N) | electrostatic stabiliser |
| Asp85A (main-N) | electrostatic stabiliser |
| Asp210A | electrostatic stabiliser |
| His212A | proton donor |
Chemical Components
proton transfer, ingold: unimolecular elimination by the conjugate base, heterolysis, intermediate collapse, overall product formed
Step 3. His232 abstracts a proton from a water which activates it to nucleophilically attack the carbon of the acyl-enzyme bond.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp83A (main-N) | electrostatic stabiliser |
| Asp85A (main-N) | electrostatic stabiliser |
| Asp210A | electrostatic stabiliser |
| His212A | proton acceptor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, intermediate formation, overall reactant used
Step 4. The oxyanion initiates an elimination which results in the cleavage if the acyl-enzyme bond and the released Ser119 can then accept a proton from His232 which returns the active site to its native state.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp83A (main-N) | electrostatic stabiliser |
| Asp85A (main-N) | electrostatic stabiliser |
| Asp210A | electrostatic stabiliser |
| Ala99A | proton acceptor |
| His212A | proton donor |
| Ala99A | nucleofuge |



 Download:
Download:  Download:
Download:  Download:
Download: