Chorismate mutase (AroQ)
Chorismate mutase (CM; EC:5.4.99.5) catalyses the reaction at the branch point of the biosynthetic pathway leading to the three aromatic amino acids, phenylalanine, tryptophan and tyrosine (chorismic acid is the last common intermediate, and CM leads to the L-phenylalanine/L-tyrosine branch). It is part of the shikimate pathway, which is present only in bacteria, fungi and plants. Members of this family, which are restricted to plants and fungi, contain a chorismate mutase domain of the AroQ class (eukaryotic type) and have an all-helical structure. The monomer consists of a catalytic and a regulatory domain covalently linked by a loop, which functions as a molecular hinge. They are monofunctional, allosteric enzymes and are subject to allosteric inhibition by tyrosine and activation by tryptophan.
Reference Protein and Structure
- Sequence
-
P32178
 (5.4.99.5)
(5.4.99.5)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Saccharomyces cerevisiae S288c (Baker's yeast)

- PDB
-
3csm
- STRUCTURE OF YEAST CHORISMATE MUTASE WITH BOUND TRP AND AN ENDOOXABICYCLIC INHIBITOR
(3.0 Å)



- Catalytic CATH Domains
-
1.10.590.10
 (see all for 3csm)
(see all for 3csm)
Enzyme Mechanism
Introduction
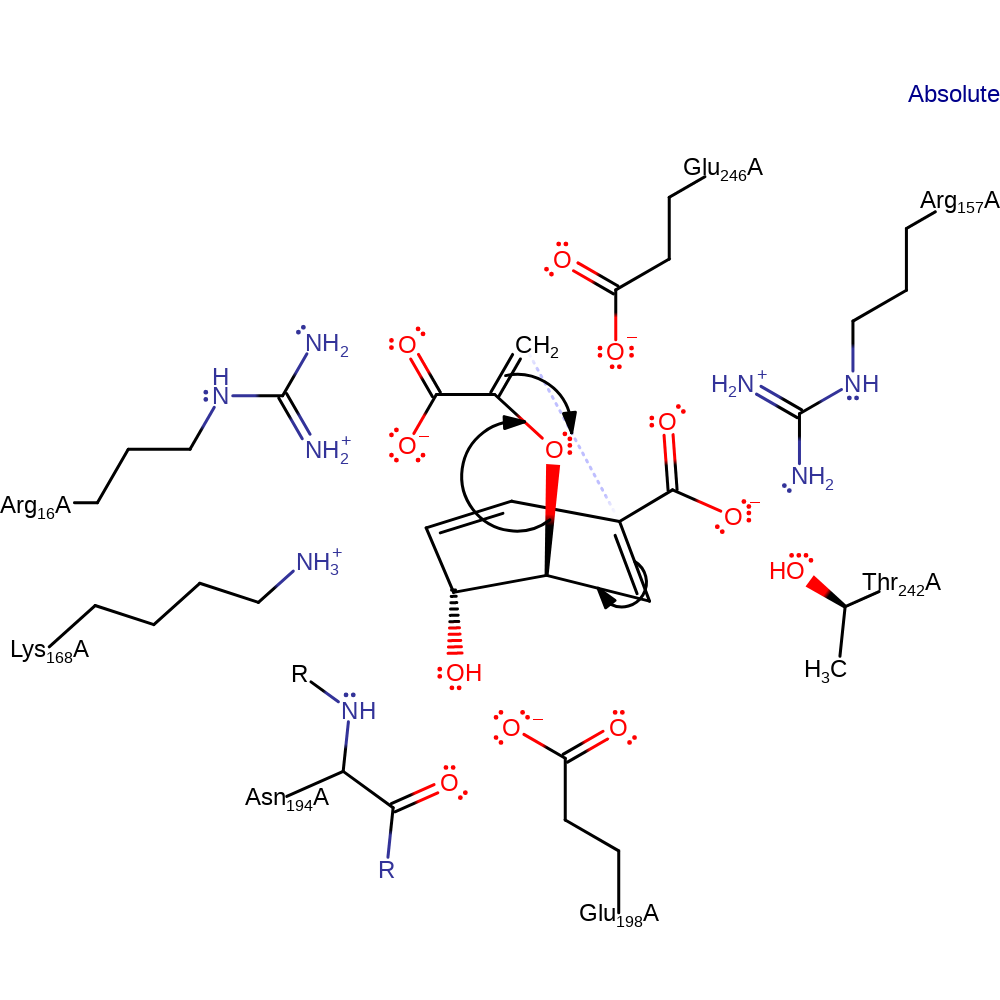
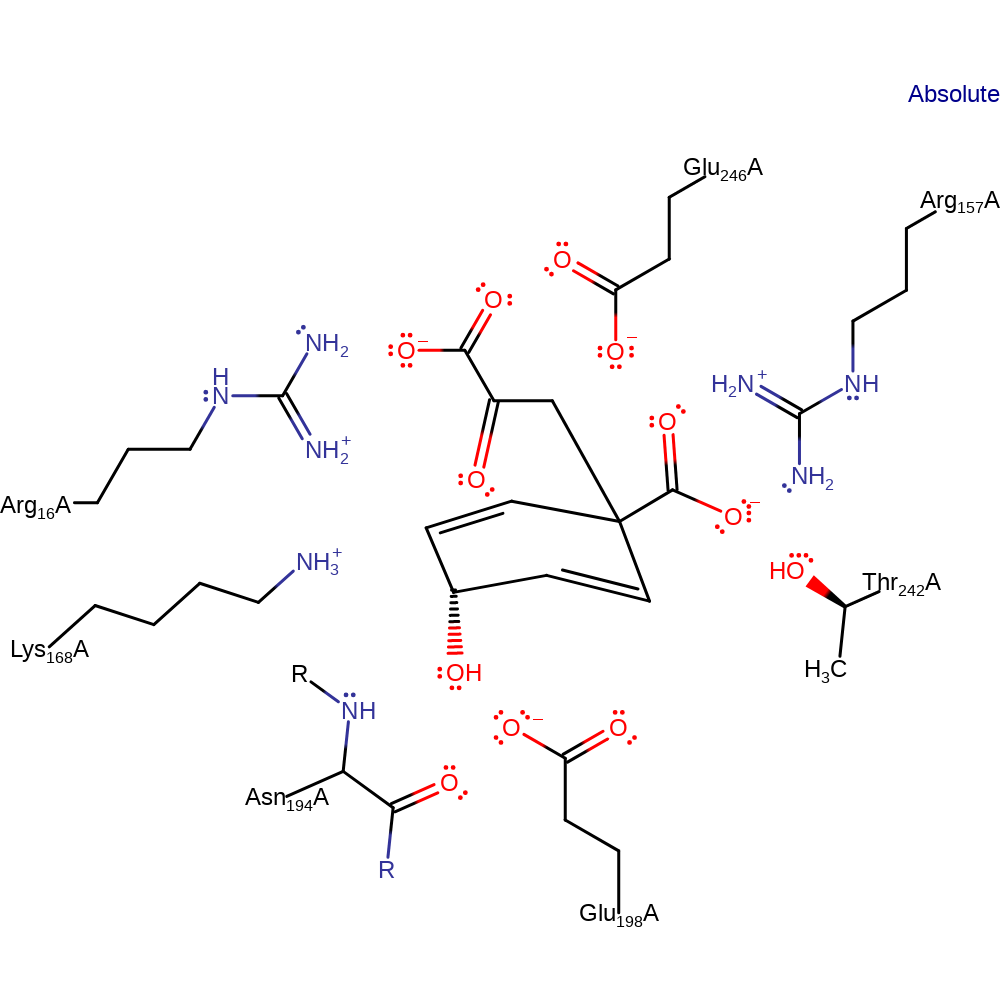
Chorismate mutase uses an extensive array of hydrogen-bonding and electrostatic interactions to bind the pseudodiaxial substrate conformer. Sterically constraining chorismate in a “near attack conformation” (NAC), in which the reacting centres are confined to contact distances. Further, ground state destabilisation through conformational compression has been shown to afford large rate accelerations for Claisen rearrangements in synthetic model systems. Polar active site residues, most notably a cationic arginine or lysine positioned next to the ether oxygen of the breaking C–O bond, stabilise the high-energy transition state electrostatically relative to the bound substrate.
Catalytic Residues Roles
| UniProt | PDB* (3csm) | ||
| Glu198, Arg16, Glu246, Arg157, Lys168, Asn194 (main-N), Thr242 | Glu198A, Arg16A, Glu246A, Arg157A, Lys168A, Asn194A (main-N), Thr242A | Forms part of the electrostatic environment that stabilises the transition state over either the substrate or product states. | hydrogen bond acceptor, transition state stabiliser |
Chemical Components
pericyclic reaction, claisen rearrangement, overall reactant used, overall product formed, rate-determining stepReferences
- Sträter N et al. (1997), Structure, 5, 1437-1452. Mechanisms of catalysis and allosteric regulation of yeast chorismate mutase from crystal structures. DOI:10.1016/s0969-2126(97)00294-3. PMID:9384560.
- Choutko A et al. (2013), Protein Sci, 22, 809-822. Exploration of swapping enzymatic function between two proteins: A simulation study of chorismate mutase and isochorismate pyruvate lyase. DOI:10.1002/pro.2264. PMID:23595942.
- Sasso S et al. (2009), EMBO J, 28, 2128-2142. Structure and function of a complex between chorismate mutase and DAHP synthase: efficiency boost for the junior partner. DOI:10.1038/emboj.2009.165. PMID:19556970.
- Kim SK et al. (2008), FEBS J, 275, 4824-4835. A comparative biochemical and structural analysis of the intracellular chorismate mutase (Rv0948c) from Mycobacterium tuberculosis H37Rv and the secreted chorismate mutase (y2828) from Yersinia pestis. DOI:10.1111/j.1742-4658.2008.06621.x. PMID:18727669.
- Lassila JK et al. (2007), Biochemistry, 46, 6883-6891. Exhaustive Mutagenesis of Six Secondary Active-Site Residues inEscherichia coliChorismate Mutase Shows the Importance of Hydrophobic Side Chains and a Helix N-Capping Position for Stability and Catalysis†. DOI:10.1021/bi700215x. PMID:17506527.
- Kim SK et al. (2006), J Bacteriol, 188, 8638-8648. Biochemical and Structural Characterization of the Secreted Chorismate Mutase (Rv1885c) from Mycobacterium tuberculosis H37Rv: an *AroQ Enzyme Not Regulated by the Aromatic Amino Acids. DOI:10.1128/jb.00441-06. PMID:17146044.
- Okvist M et al. (2006), J Mol Biol, 357, 1483-1499. 1.6Å Crystal Structure of the Secreted Chorismate Mutase from Mycobacterium tuberculosis: Novel Fold Topology Revealed. DOI:10.1016/j.jmb.2006.01.069. PMID:16499927.
- Qamra R et al. (2006), Biochemistry, 45, 6997-7005. The 2.15 Å Crystal Structure ofMycobacterium tuberculosisChorismate Mutase Reveals an Unexpected Gene Duplication and Suggests a Role in Host−Pathogen Interactions†. DOI:10.1021/bi0606445. PMID:16752890.
- Zhang X et al. (2005), Biochemistry, 44, 10443-10448. A Definitive Mechanism for Chorismate Mutase†. DOI:10.1021/bi050886p. PMID:16060652.
- Hur S et al. (2003), J Am Chem Soc, 125, 5964-5972. Comparison of Formation of Reactive Conformers (NACs) for the Claisen Rearrangement of Chorismate to Prephenate in Water and in theE.coliMutase: The Efficiency of the Enzyme Catalysis. DOI:10.1021/ja0210648. PMID:12733937.
- Helmstaedt K et al. (2002), Proc Natl Acad Sci U S A, 99, 6631-6636. Refined molecular hinge between allosteric and catalytic domain determines allosteric regulation and stability of fungal chorismate mutase. DOI:10.1073/pnas.092130899. PMID:11997452.
- Hur S et al. (2002), Proc Natl Acad Sci U S A, 99, 1176-1181. The mechanism of catalysis of the chorismate to prephenate reaction by the Escherichia coli mutase enzyme. DOI:10.1073/pnas.022628599. PMID:11818529.
- Guo H et al. (2001), Proc Natl Acad Sci U S A, 98, 9032-9037. Substrate conformational transitions in the active site of chorismate mutase: Their role in the catalytic mechanism. DOI:10.1073/pnas.141230998. PMID:11481470.
- Khanjin NA et al. (1999), J Am Chem Soc, 121, 11831-11846. Mechanism of Chorismate Mutase: Contribution of Conformational Restriction to Catalysis in the Claisen Rearrangement. DOI:10.1021/ja992453d.
- Ma J et al. (1998), Proc Natl Acad Sci U S A, 95, 14640-14645. Yeast chorismate mutase in the R state: Simulations of the active site. DOI:10.1073/pnas.95.25.14640. PMID:9843942.
- Lee AY et al. (1998), Biochemistry, 37, 9052-9057. Thermodynamics of a Transition State Analogue Inhibitor Binding toEscherichia coliChorismate Mutase: Probing the Charge State of an Active Site Residue and Its Role in Inhibitor Binding and Catalysis†,‡. DOI:10.1021/bi980217u. PMID:9636050.
- Christendat D et al. (1998), Biochemistry, 37, 15703-15712. Use of Site-Directed Mutagenesis To Identify Residues Specific for Each Reaction Catalyzed by Chorismate Mutase−Prephenate Dehydrogenase fromEscherichia coli†. DOI:10.1021/bi981412b. PMID:9843375.
- Schnappauf G et al. (1998), J Biol Chem, 273, 17012-17017. Tyrosine and tryptophan act through the same binding site at the dimer interface of yeast chorismate mutase. PMID:9642265.
- Lin SL et al. (1997), J Mol Biol, 271, 838-845. Investigation of the enzymatic mechanism of the yeast chorismate mutase by docking a transition state analog. DOI:10.1006/jmbi.1997.1168. PMID:9299331.
- Liu DR et al. (1996), J Am Chem Soc, 118, 1789-1790. Analysis of Active Site Residues inEscherichia coliChorismate Mutase by Site-Directed Mutagenesis. DOI:10.1021/ja953151o.
- Strater N et al. (1996), Proc Natl Acad Sci U S A, 93, 3330-3334. Crystal structure of the T state of allosteric yeast chorismate mutase and comparison with the R state. PMID:8622937.
- Xue Y et al. (1995), Proc Natl Acad Sci U S A, 92, 10595-10598. Location of the active site of allosteric chorismate mutase from Saccharomyces cerevisiae, and comments on the catalytic and regulatory mechanisms. DOI:10.1073/pnas.92.23.10595. PMID:7479847.
- Lee AY et al. (1995), J Am Chem Soc, 117, 3627-3628. Atomic structure of the buried catalytic pocket of Escherichia coli chorismate mutase. DOI:10.1021/ja00117a038.
Catalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu198A | hydrogen bond acceptor |
| Arg16A | hydrogen bond donor |
| Lys168A | hydrogen bond donor |
| Glu246A | hydrogen bond donor |
| Arg157A | hydrogen bond donor |
| Arg16A | transition state stabiliser |
| Arg157A | transition state stabiliser |
| Lys168A | transition state stabiliser |
| Asn194A (main-N) | transition state stabiliser |
| Glu198A | transition state stabiliser |
| Thr242A | transition state stabiliser |
| Glu246A | transition state stabiliser |


 Download:
Download: