Ribulose-bisphosphate carboxylase (type II)
Ribulose-bisphosphate carboxylase/oxygenase (RuBisCO) is the enzyme responsible for the initial step of carbon fixation during photosynthesis. RuBisCO carboxylates one molecule of ribulose-bisphosphate (RuBP), yielding two molecules of 3-phosphoglycerate(PGA). The latter is subsequently used partly to regenerate RuBP in the Calvin cycle and partly to build up sugar compounds. RuBisCO also acts as an oxygenase with RuBP as substrate. The oxygenation of RuBP initiates photorespiration, a pathway in which energy initially accumulated through the light reactions is lost as heat, with no apparent benefit for the plant. In higher plants and the majority of photosynthetic microorganisms, RuBP-carboxylase is composed of eight large and eight small subunits, forming a hexadecameric structure, L8S8. In contrast, in some photosynthetic bacteria, RuBisCO is a dimer of only large subunits.
Reference Protein and Structure
- Sequence
-
P04718
 (4.1.1.39)
(4.1.1.39)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Rhodospirillum rubrum (Bacteria)

- PDB
-
1rba
- SUBSTITUTION OF ASP193 TO ASN AT THE ACTIVE SITE OF RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE RESULTS IN CONFORMATIONAL CHANGES
(2.6 Å)



- Catalytic CATH Domains
-
3.20.20.110
 (see all for 1rba)
(see all for 1rba)
- Cofactors
- Magnesium(2+) (1)
Enzyme Reaction (EC:4.1.1.39)
Enzyme Mechanism
Introduction
Rubisco has to be activated for reaction. Activation is achieved by carbamylation of a lysine residue(Lys191). This reaction creates a binding site for a Mg(II) ion that stabilises the carbamate by monodentate and completes the active site of the enzyme. After activation, RuBP binds to the enzyme, O2 and O3 become coordinated to Mg(II) ions, displacing 2 water molecules. The coodination of O2 and O3 by Mg(II) ion increases acidity of the protons attached to C3 and O3, thus assisting their subsequent removal. The first step of the reaction is the removal of the proton at the C3 carbon atom of RUBP by carbamate. The combined effect of the Mg2+ polarising the C2 carbonyl and the proximity of a carbamino oxygen positioned to accept the proton favours the formation of the C2-C3 enediolate. The enediol intermediate is further stabilised by protonation of the C2 oxygen by the protonated carbamate. The fact that C3 proton is not conserved in upper 3-P-lgycerate is explained by a proton transfer from C3 to the carbamate and then to O2 which is then abstracted by Lys166. An enediolate is created in which O2 is unprotonated and O3 is protonated and again, carbamate facilitates the transfer of proton from O3 to O2. The next step is the addition of CO2 to the enediolate intermediate. The precise details of the reaction of the enediolate with CO2 and water are unclear. Both a sequential, two step reaction and a concerted mechanism are plausible. In the two step sequential mechanism, the electrons from the enediolate oxygen at C3 forms a double bond, permitting attack of C2 on CO2 to form an intermediate with a keto group at C3. Then, a water molecule, activated by His287, nucleophilically attack the C3 keto group. The alternative mechanism is a concerted one in which attack of water at C3 occurs in the same transition state as attack on CO2. In both case, Lys329 facilitates the addition of CO2 by polarising the 2 oxygen of CO2 and in the former case, it also stabilises the transition state of the carboxylate intermediate. Once the six-carbon hydrated ketone intermediate is formed, C2-C3 bond is cleaved. Cleavage of C2-C3 bond is facilitated by proton removal from OH group at C3. Carbamate on Lys191, again, acts as a base here. Following cleavage of the bond between C2 and C3, one molecule of 3-PGA is immediately yielded from carbon atoms C3 to C5, whereas the second molecule of D-3-PGA is produced by stereospecific protonation of the carbanion derived from C1 and C2.Protonated Lys166 is ideally positioned to act as the acid for this step. The rotation of C1 and C2 leading to aci-carboxylate formation brings C2 in contact with Lys166 and facilitate the protonation. Side chain carbonyl oxygen of Asn192 forms a hydrogen bond to carbamate NH to increase the positive charge of the nitrogen, enhancing the effectiveness of carbamate to act as a base. Based on site mutagenesis, His321 is also found to be stabilising the transition states of the reaction.
Catalytic Residues Roles
| UniProt | PDB* (1rba) | ||
| His287 | His287A | It acts as a general base for water attack at C-3. | activator, increase nucleophilicity, proton acceptor |
| Asn192 | Asn192A | Its carbonyl oxygen accepts H-bond from carbamate NH, increasing the positive charge on this nitrogen. | electrostatic stabiliser |
| Lys191 | Lys191A | It reacts with CO2 to form carbamate which provides monodentate ligand to Mg(II) to complete the active site. The non-metal-coordinated oxygen of the carbamate serves as the general base for removing the C-3 proton of RuBP, transferring it via the O-2 of RuBP to Lys166. It facilitates proton transfer between O-3 and O-2 of the enediolate and serves as the general base to remove the non-metal-coordinated hydroxyl proton at C-3 of the gem-diol form of 2C3KABP to initiate C-2/C-3 bond cleavage. | electron pair donor, nucleophile, metal ligand |
| Asp193, Glu194, Lys191 | Asn193A, Glu194A, Lys191A | Coordinate the magnesium ion. | metal ligand |
| His321 | His321A | It maybe involved in transition state stabilisation. | electrostatic stabiliser |
| Lys166 | Lys166A | It accepts proton after enolisation and it protonates aci-carboxylate in last step | proton acceptor, electrostatic stabiliser, proton donor |
Chemical Components
bimolecular nucleophilic addition, tautomerisation (not keto-enol), coordination to a metal ion, overall reactant used, assisted keto-enol tautomerisation, proton transfer, bimolecular electrophilic addition, overall product formed, heterolysis, native state of enzyme is not regeneratedReferences
- Andersson I (1996), J Mol Biol, 259, 160-174. Large Structures at High Resolution: The 1.6 Å Crystal Structure of Spinach Ribulose-1,5- Bisphosphate Carboxylase/Oxygenase Complexed with 2-Carboxyarabinitol Bisphosphate. DOI:10.1006/jmbi.1996.0310. PMID:8648644.
- Cleland WW et al. (1998), Chem Rev, 98, 549-562. Mechanism of Rubisco: The Carbamate as General Base✗. DOI:10.1021/cr970010r. PMID:11848907.
- Harpel MR et al. (1996), Biochemistry, 35, 13865-13870. Facilitation of the Terminal Proton Transfer Reaction of Ribulose 1,5-Bisphosphate Carboxylase/Oxygenase by Active-Site Lys166†. DOI:10.1021/bi962184t. PMID:8909282.
- Gutteridge S et al. (1995), Plant Cell, 7, 809-819. Rubisco Synthesis, Assembly, Mechanism, and Regulation. DOI:10.1105/tpc.7.7.809. PMID:12242387.
- Harpel MR et al. (1991), J Biol Chem, 266, 24734-24740. Functional analysis of the putative catalytic bases His-321 and Ser-368 of Rhodospirillum rubrum ribulose bisphosphate carboxylase/oxygenase by site-directed mutagenesis. PMID:1761567.
- Hartman FC et al. (1989), J Biol Chem, 264, 11784-11789. Examination of the function of active site lysine 329 of ribulose-bisphosphate carboxylase/oxygenase as revealed by the proton exchange reaction. PMID:2545684.
- Soper TS et al. (1988), Protein Eng, 2, 39-44. Essentiality of Lys-329 of ribulose-1,5-bisphosphate carboxylase/oxygenase from Rhodospirillum rubrum as demonstrated by site-directed mutagenesis. DOI:10.1093/protein/2.1.39. PMID:3151016.
- Lorimer GH et al. (1988), J Biol Chem, 263, 6468-6471. Evidence supporting lysine 166 of Rhodospirillum rubrum ribulosebisphosphate carboxylase as the essential base which initiates catalysis. PMID:3129424.
- Estelle M et al. (1985), J Biol Chem, 260, 9523-9526. Site-specific mutagenesis of ribulose-1,5-bisphosphate carboxylase/oxygenase. Evidence that carbamate formation at Lys 191 is required for catalytic activity. PMID:2991249.
- Lorimer GH (1981), Biochemistry, 20, 1236-1240. Ribulosebisphosphate carboxylase: amino acid sequence of a peptide bearing the activator carbon dioxide. DOI:10.1021/bi00508a028. PMID:6784749.
- Lorimer GH et al. (1980), Biochemistry, 19, 5321-5328. Carbamate formation on the .epsilon.-amino group of a lysyl residue as the basis for the activation of ribulosebisphosphate carboxylase by carbon dioxide and magnesium(2+). DOI:10.1021/bi00564a027. PMID:6778504.

Step 1. Lys191 carries out a nucleophilic attack on a CO2 molecule. This will produce an intermediate which will become a carbamate which will coordinate the magnesium ion in the later stages of catalysis.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys191A | nucleophile |
Chemical Components
ingold: bimolecular nucleophilic addition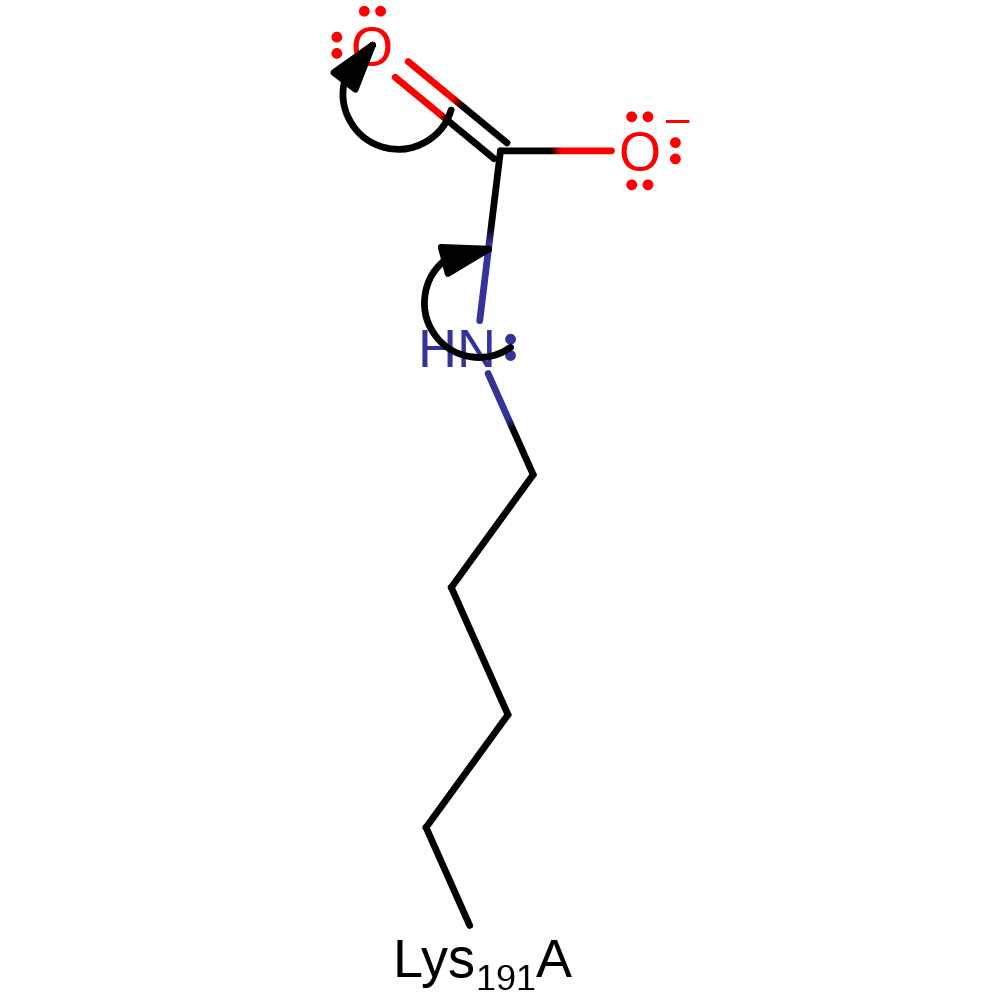
Step 2. The intermediate tautomerizes into the carbamate. Magnesium and RUBP binding then occur sequentially.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys191A | electron pair donor |
Chemical Components
tautomerisation (not keto-enol), coordination to a metal ion
Step 3. The uncoordinated oxyanion of the carbamate deprotonates C3, causing a keto-enol tautomerization. The enolate is stabilized by the magnesium ion, Lys166, Lys329 and His321.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn192A | electrostatic stabiliser |
| His321A | electrostatic stabiliser |
| Lys166A | electrostatic stabiliser |
| LysNone(329)A | electrostatic stabiliser |
| Lys191A | metal ligand |
| Asn193A | metal ligand |
| Glu194A | metal ligand |
Chemical Components
overall reactant used, assisted keto-enol tautomerisation, proton transfer
Step 4. Water attacks C3 of the C=C bond and C2 attacks a CO2 molecule. Lys166 abstracts a proton from the C3 hydroxyl and His287 activates the water molecule.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys166A | electrostatic stabiliser |
| His321A | electrostatic stabiliser |
| LysNone(329)A | electrostatic stabiliser |
| Lys191A | metal ligand |
| Asn193A | metal ligand |
| Glu194A | metal ligand |
| His287A | activator, increase nucleophilicity, proton acceptor |
| Lys166A | proton acceptor |
Chemical Components
ingold: bimolecular electrophilic addition, ingold: bimolecular nucleophilic addition, proton transfer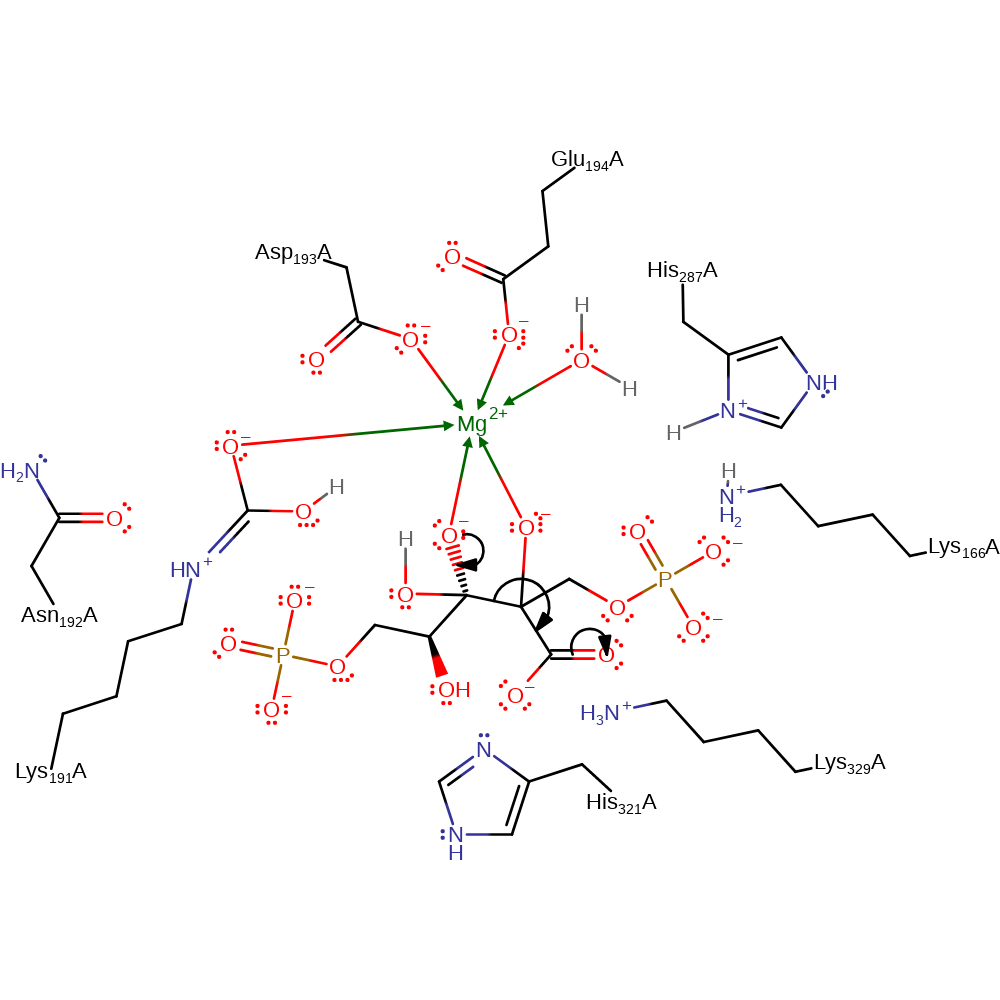
Step 5. The C3-C2 bond is cleaved forming one of the 3-phosphoglycerate products and an enolate of another 3PG molecule.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys166A | electrostatic stabiliser |
| His321A | electrostatic stabiliser |
| LysNone(329)A | electrostatic stabiliser |
| Lys191A | metal ligand |
| Asn193A | metal ligand |
| Glu194A | metal ligand |
Chemical Components
overall product formed, heterolysis
Step 6. The enolate intermediate tautomerizes into 3PG, this is assisted by protonation from Lys166 and the Lys191 carbamate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys166A | electrostatic stabiliser |
| Asn192A | electrostatic stabiliser |
| His321A | electrostatic stabiliser |
| LysNone(329)A | electrostatic stabiliser |
| Lys191A | metal ligand |
| Asn193A | metal ligand |
| Glu194A | metal ligand |
| Lys166A | proton donor |





 Download:
Download: