Aspartate transaminase
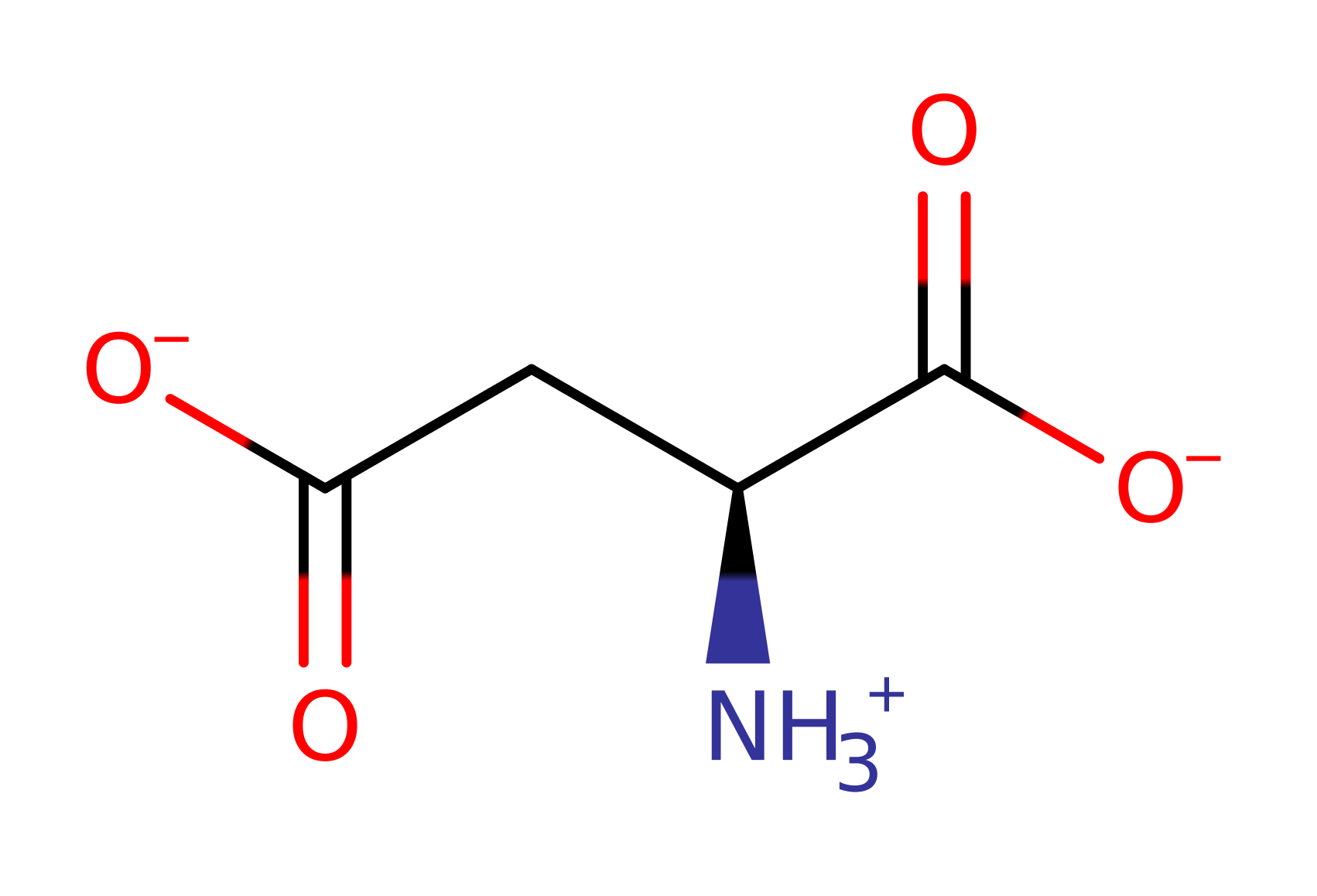
Aspartyl transferase is able to catalyse the PLP dependent transamination reaction between aspartate and 2-oxoglutarate, forming oxaloacetate and glutamate. It is part of the family of PLP dependent amino acid transferase enzymes which have high sequence homology and identical active site organisation, with the only difference being in the amino acid and ketoacid substrates. The enzymes all play key roles in the catabolism of amino acids as the products feed into the Krebs cycle and the Urea cycle.
Reference Protein and Structure
- Sequence
-
P00509
 (2.6.1.1)
(2.6.1.1)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Escherichia coli K-12 (Bacteria)

- PDB
-
1aam
- THE STRUCTURAL BASIS FOR THE ALTERED SUBSTRATE SPECIFICITY OF THE R292D ACTIVE SITE MUTANT OF ASPARTATE AMINOTRANSFERASE FROM E. COLI
(2.8 Å)



- Catalytic CATH Domains
-
3.40.640.10
 (see all for 1aam)
(see all for 1aam)
- Cofactors
- Pyridoxal 5'-phosphate(2-) (1)
Enzyme Reaction (EC:2.6.1.1)
Enzyme Mechanism
Introduction
The reaction follows a ping-pong mechanism.
In the enzyme, PLP is anchored to the protein at Lys258 with its pyridoxal moiety in the reactive bipolar ionic form. In the first step, the cofactor PLP is transferred from Lys 258 to the aspartate alpha amino group. The alpha-amino group of the aspartate attacks the PLP C4' from the front side in a direction perpendicular to the plane of the pyridine ring. The attraction of opposite charges on the substrate N atom and O3' of the coenzyme brings about a 90 degrees rotation of the 2 C-N bonds around C4-C4' which brings Lys258 behind the plane of the pyridine ring and hence Lys258 can be released. Trp 140 sterically constraining the PLP to allow the attack of aspartate.
The deprotonation of the alpha-carbon of aspartate gives rise to the quinonoid intermediate, which is stabilised by the electron link of the protonated pyridine ring. Asp223 facilitates the deprotonation by stabilising the positive charge at N1 of PLP with a salt bridge and hence enhancing the electron withdrawing capacity of the amino acid substrate. Lys 258 acts as a base here. Lys258 then protonates C4' from the si side, giving rise to keimine intermediate. It then deprotonates a water molecule to allow its nucleophilic attack on the alpha-carbon. The tetrahedral intermediate dissociates into PMP and oxo-acid product. Subsequent reversal of the reaction steps described occurs except with the carboxyl group of the alpha keto-glutarate acting as the nucleophile leading to the formation of glutamate and the completion of the reaction cycle.
Catalytic Residues Roles
| UniProt | PDB* (1aam) | ||
| Asp211 | Asp223(211)A | It forms a salt bridge with N1 of pyridine ring and enhance the electron withdrawing capacity of the pyridine ring to facilitate the removal of the alpha-proton of the amino acid substrate. It also acts as acid base to facilitate deprotonation of the aspartate's alpha amino group to allow it to act as a nucleophile and attack the PLP. | proton shuttle (general acid/base) |
| Lys246 | Lys258(246)A | Binds to PLP cofactor. Also acts as acid base for the conversion of the aldimine to the ketimine forms of the PLP-aspartate intermediate. | proton shuttle (general acid/base) |
| Trp130 | Trp142(130)A | Sterically constrains the PLP cofactor in order to make nucleophilic attack by the aspartate on PLP more favourable. | steric role |
Chemical Components
References
- Yano T et al. (1992), Biochemistry, 31, 5878-5887. Role of Asp222 in the catalytic mechanism of Escherichia coli aspartate aminotransferase: the amino acid residue which enhances the function of the enzyme-bound coenzyme pyridoxal 5'-phosphate. DOI:10.1021/bi00140a025. PMID:1610831.
- Smith DL et al. (1989), Biochemistry, 28, 8161-8167. 2.8-.ANG.-resolution crystal structure of an active-site mutant of aspartate aminotransferase from Escherichia coli. DOI:10.1021/bi00446a030. PMID:2513875.
- Kirsch JF et al. (1984), J Mol Biol, 174, 497-525. Mechanism of action of aspartate aminotransferase proposed on the basis of its spatial structure. DOI:10.1016/0022-2836(84)90333-4. PMID:6143829.
Catalytic Residues Roles
| Residue | Roles |
|---|---|
| Trp142(130)A | steric role |
| Asp223(211)A | proton shuttle (general acid/base) |
| Lys258(246)A | proton shuttle (general acid/base) |