D-aminoacyl-tRNA deacylase
YihZ, a D-Tyr-tRNA(Tyr) deacylase from Haemophilus influenzae is an 'editing enzyme'. It removes D-tyrosine and other D-amino acids from charged tRNAs, thereby preventing incorrect incorporation of D-amino acids in proteins. This is important, as the presence of D-amino acids can impair correct folding of proteins. Any incorrectly charged tRNA must be removed, and YihZ catalyses the hydrolysis of the ester link between D-Tyr and tRNA. This enzyme exhibits broad specificity towards D-amino acids, but it is inactive toward L-aminoacylated tRNAs and N-blocked D-aminoacylated tRNAs.
Reference Protein and Structure
- Sequence
-
P44814
 (3.1.1.96)
(3.1.1.96)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Haemophilus influenzae Rd KW20 (Bacteria)

- PDB
-
1j7g
- Structure of YihZ from Haemophilus influenzae (HI0670), a D-Tyr-tRNA(Tyr) deacylase
(1.64 Å)



- Catalytic CATH Domains
-
3.50.80.10
 (see all for 1j7g)
(see all for 1j7g)
Enzyme Reaction (EC:3.1.1.96)
Enzyme Mechanism
Introduction
Thr 80 is substrate assisted, its deprotonation by the deprotonated amino nitrogen atom of D-Tyr activates it for nucleophilic attack. The hydroxyl group of Thr 80 nucleophilically attacks the carbonyl carbon of D-Tyr, forming a negatively charged, tetrahedral intermediate. This intermediate is stabilised by the oxyanion hole which is composed of the main chain amides of Thr 80 and Phe 79, and the side chain of Gln 78. As the carbonyl is reformed, the ester link is broken, and the leaving group tRNA molecule is protonated by the substrate D-Tyr. D-Tyr then assists again by deprotonating a water molecule, activating it for nucleophilic attack on the D-Tyr carbonyl carbon. Nucleophilic attack results in another negatively charged, tetrahedral intermediate which is again stabilised by the oxyanion hole. As the carbonyl is reformed, the link to Thr 80 is broken, and Thr 80 is re-protonated by the D-Tyr amino nitrogen.
Catalytic Residues Roles
| UniProt | PDB* (1j7g) | ||
| Phe79 (main-N) | Phe79A (main-N) | The main chain amide of Phe 79 forms part of the oxyanion hole, serving to stabilise the negatively charged intermediate. | electrostatic stabiliser |
| Gln78 | Gln78A | The side chain amide of Gln 78 forms part of the oxyanion hole, serving to stabilise the negatively charged intermediate. | electrostatic stabiliser |
| Thr80 | Thr80A | The hydroxyl of Thr 80 is activated, and then nucleophilically attacks the carbonyl carbon of D-Tyr. The main chain amide of Thr 80 forms part of the oxyanion hole, and serves to stabilise the negatively charged intermediate. | nucleofuge, nucleophile, proton acceptor, proton donor |
Chemical Components
proton transfer, bimolecular nucleophilic addition, enzyme-substrate complex formation, intermediate formation, overall reactant used, unimolecular elimination by the conjugate base, heterolysis, intermediate collapse, overall product formed, native state of enzyme regeneratedReferences
- Lim K et al. (2003), J Biol Chem, 278, 13496-13502. A Catalytic Mechanism for D-Tyr-tRNATyrDeacylase Based on the Crystal Structure of Hemophilus influenzae HI0670. DOI:10.1074/jbc.m213150200. PMID:12571243.
- Bhatt TK et al. (2010), J Biol Chem, 285, 5917-5930. Ligand-bound structures provide atomic snapshots for the catalytic mechanism of D-amino acid deacylase. DOI:10.1074/jbc.M109.038562. PMID:20007323.
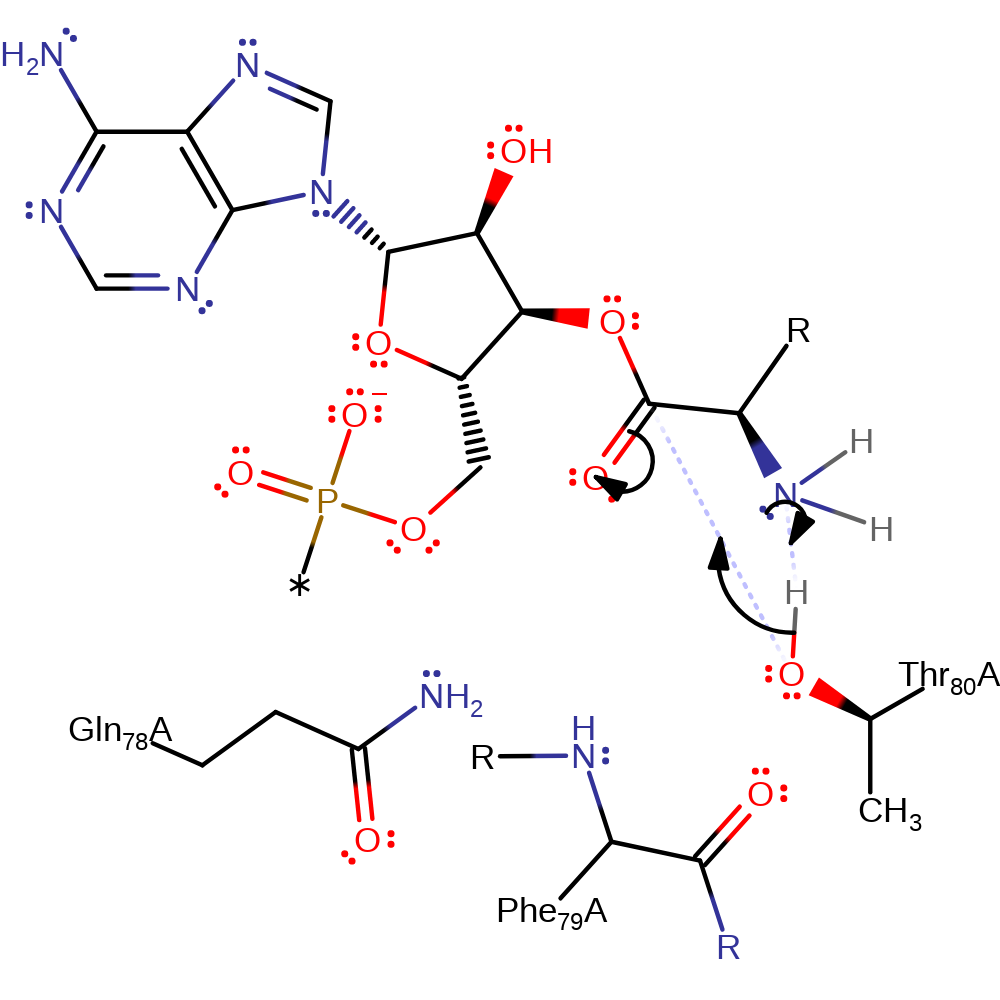
Step 1. The amino nitrogen of D-Tyr deprotonates Thr80 which can then nucleophilically attack the carbon of the carbonyl.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Phe79A (main-N) | electrostatic stabiliser |
| Gln78A | electrostatic stabiliser |
| Thr80A | proton donor, nucleophile |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, enzyme-substrate complex formation, intermediate formation, overall reactant used
Step 2. The amino group of D-tyr protonates the oxygen of the ester bond which initiates an elimination from the oxyanion and results in the cleavage of the t-RNA-Tyr bond.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gln78A | electrostatic stabiliser |
| Phe79A (main-N) | electrostatic stabiliser |
Chemical Components
proton transfer, ingold: unimolecular elimination by the conjugate base, heterolysis, intermediate collapse, overall product formed
Step 3. D-Tyr's amino group deprotonates a water which activates it to nucleophilically attack the carbonyl carbon.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gln78A | electrostatic stabiliser |
| Phe79A (main-N) | electrostatic stabiliser |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, intermediate formation, overall reactant used
Step 4. The amino group protonates the oxygen of Thr80 which initiates another elimination from the oxyanion which results in the cleavage of the acyl-enzyme bond.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gln78A | electrostatic stabiliser |
| Phe79A (main-N) | electrostatic stabiliser |
| Thr80A | proton acceptor, nucleofuge |
Chemical Components
proton transfer, ingold: unimolecular elimination by the conjugate base, heterolysis, intermediate collapse, native state of enzyme regenerated, overall product formedIntroduction
Some experiments have shown that mutation of Thr80 does not reduce the activity enough of the enzyme and in fact the deacylation still occurs with a mutated Threonine. Therefore this suggests that it is not involved in the catalytic mechanism. Instead an RNA-assisted catalytic mechanism implicating the role of 2′-OH in activating a water molecule for catalysis has been proposed. The 2′-OH of the terminal ribose would activate a water molecule, which in turn makes a nucleophilic attack on the carbonyl carbon of the substrate. The resultant tetrahedral transition state would be stabilized by the oxyanion hole formed by main chain nitrogen atoms of Phe 79 and Thr 80 and Gln 78. It would then result in the subsequent cleavage of the ester bond between the D-Tyr and the tRNA
Catalytic Residues Roles
| UniProt | PDB* (1j7g) | ||
| Thr80 (main-N), Phe79 (main-N), Gln78 | Thr80A (main-N), Phe79A (main-N), Gln78A | Form the oxyanion hole and stabilise the negative charge of the tetrahedral transition state through hydrogen bonding. | electrostatic stabiliser |
Chemical Components
proton transfer, bimolecular nucleophilic addition, intermediate formation, overall reactant used, unimolecular elimination by the conjugate base, intermediate collapse, overall product formedReferences
- Ahmad S et al. (2013), Elife, 2, e01519-. Mechanism of chiral proofreading during translation of the genetic code. DOI:10.7554/eLife.01519. PMID:24302572.

Step 1. The 2' OH of the ribose abstracts a proton from a water which activates it to nucleophilically attack the carbonyl carbon.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gln78A | electrostatic stabiliser |
| Phe79A (main-N) | electrostatic stabiliser |
| Thr80A (main-N) | electrostatic stabiliser |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, intermediate formation, overall reactant used
Step 2. The protonation of the ester oxygen initiates an elimination from the oxyanion which results in the cleavage of the ester bond which releases D-Tyr from tRNA.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gln78A | electrostatic stabiliser |
| Phe79A (main-N) | electrostatic stabiliser |
| Thr80A (main-N) | electrostatic stabiliser |





 Download:
Download:  Download:
Download: