DNA topoisomerase (ATP-hydrolysing)
E. coli DNA gyrase is a member of the family of type II topoisomerases, which change the topology of DNA by cleaving both the phosphodiester backbones of the double-stranded DNA, transporting another double-stranded segement through the enzyme bridged gap, and then religating the DNA. Therefore, E. coli DNA gyrase catalyses the relaxation of negatively supercoiled DNA, and is uniquely also able to introduce negative supercoils into DNA at the expense of ATP hydrolysis. By introducing a transient break into the DNA substrate, pasing another piece of DNA though the gap and releasing it, the enzyme relieves topological contraints caused by replication and transcription complexes moving along the DNA. The enzyme consists of 2 GyrA and 2 GyrB subunits. The catalytic domain is found on the GyrA subunit, whilst the ATP hydrolysis site is found on GyrB.
Reference Protein and Structure
- Sequences
-
P0AES4
 (5.6.2.2)
(5.6.2.2)
P0AES7 (5.6.2.2)
(5.6.2.2)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Escherichia coli K-12 (Bacteria)

- PDB
-
1ab4
- 59KDA FRAGMENT OF GYRASE A FROM E. COLI
(2.8 Å)



- Catalytic CATH Domains
-
3.90.199.10
 (see all for 1ab4)
(see all for 1ab4)
- Cofactors
- Magnesium(2+) (2)
Enzyme Reaction (EC:5.6.2.2)
Enzyme Mechanism
Introduction
The DNA gyrase enzyme catalyses the cleavage of and religation of DNA. The active site Tyr 122 is polarised by the magnesium ion, making it more susceptible to deprotonation by the his 78 general base of GyrA. Tyr 122 nucleophilically attacks the scissile phosphate group, which forms a negatively charged pentacovalent phosphate intermediate. This intermediate is stabilised by both magnesium ions and also by the positively charged side chain of Arg 32 on the GyrA subunit. The 3' bridging O atom is stabilised by Mg2+ to make to 3' oxyanion a better leaving group and to ensure the 3'-hydroxyl group is produced. The unstable 3'-oxyanion group is protonated by an unidentified acid (possibly conserved Lys 449 of GyrB).
Catalytic Residues Roles
| UniProt | PDB* (1ab4) | ||
| His78 | His78(49)A | The His 78 acts as a general base, deprotonating the nucleophilic Tyr 122 residue. | proton acceptor |
| Arg32 | Arg32(3)A | The side chain of Arg 32 stabilises the transition state through hydrogen bonding. | electrostatic stabiliser |
| Tyr122 | Tyr122(93)A(AA) | Tyr 122 is deprotonated by the general base His 78, and is also polarised by a co-ordinated magnesium ion. This activates Tyr 122 to nucleophilic attack of the substrate scissile phosphate group. | nucleofuge, nucleophile, metal ligand, proton acceptor, proton donor |
| Asp498, Asp500, Asp502, Glu424 | Not found, Not found, Not found, Not found | Coordinate the Mg ions. | metal ligand |
Chemical Components
proton transfer, bimolecular nucleophilic addition, intermediate formation, intramolecular elimination, proton relay, intermediate terminated, native state of enzyme regeneratedReferences
- Noble CG et al. (2002), J Mol Biol, 318, 361-371. The Role of GyrB in the DNA Cleavage-religation Reaction of DNA Gyrase: A Proposed Two Metal-ion Mechanism. DOI:10.1016/s0022-2836(02)00049-9. PMID:12051843.
- Hanaoka K et al. (2014), J Biomol Struct Dyn, 32, 1759-1765. Substrate-mediated proton relay mechanism for the religation reaction in topoisomerase II. DOI:10.1080/07391102.2013.834848. PMID:24047515.
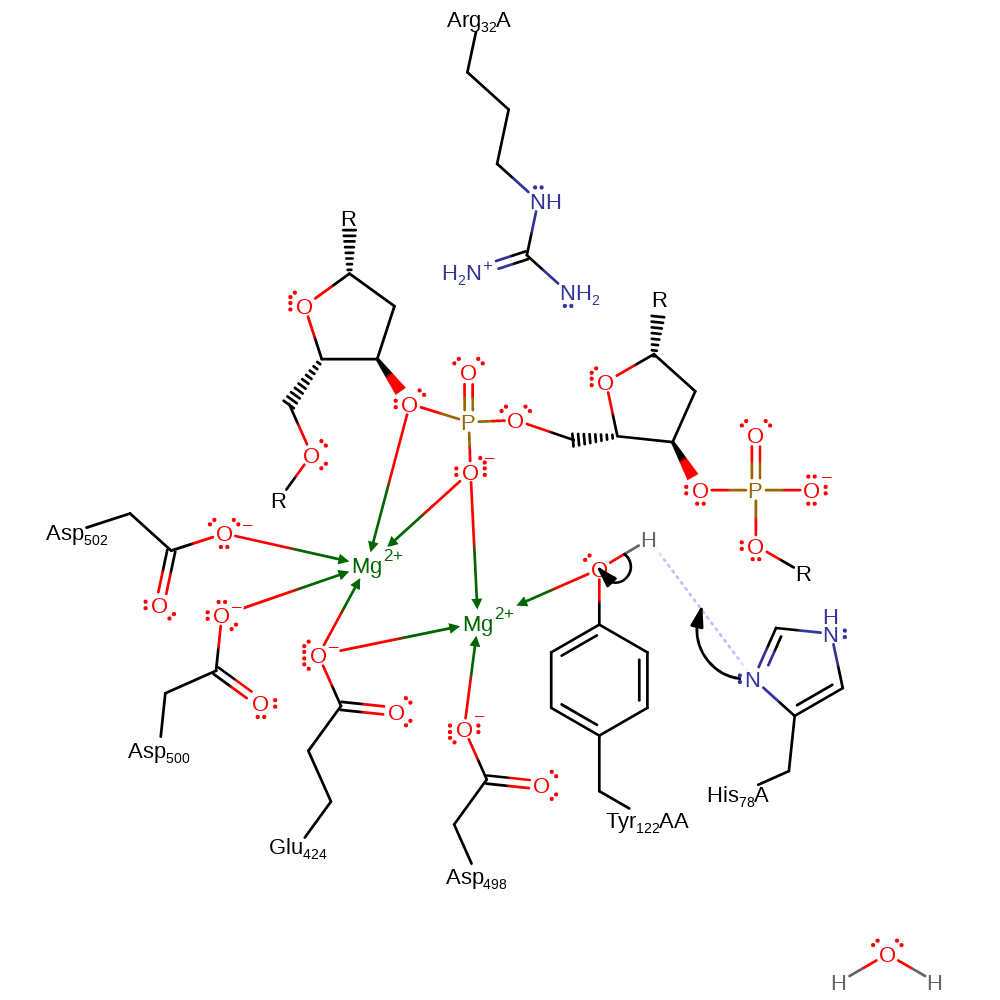
Step 1. His 78 deprotonates Tyr 122, activating the phenol group. The oxygen of the phenol is coordinated to the second Mg ion which stabilizes the conjugate base.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu424 | metal ligand |
| Asp498 | metal ligand |
| Asp500 | metal ligand |
| Asp502 | metal ligand |
| Tyr122(93)A(AA) | metal ligand |
| His78(49)A | proton acceptor |
| Tyr122(93)A(AA) | proton donor |
Chemical Components
proton transfer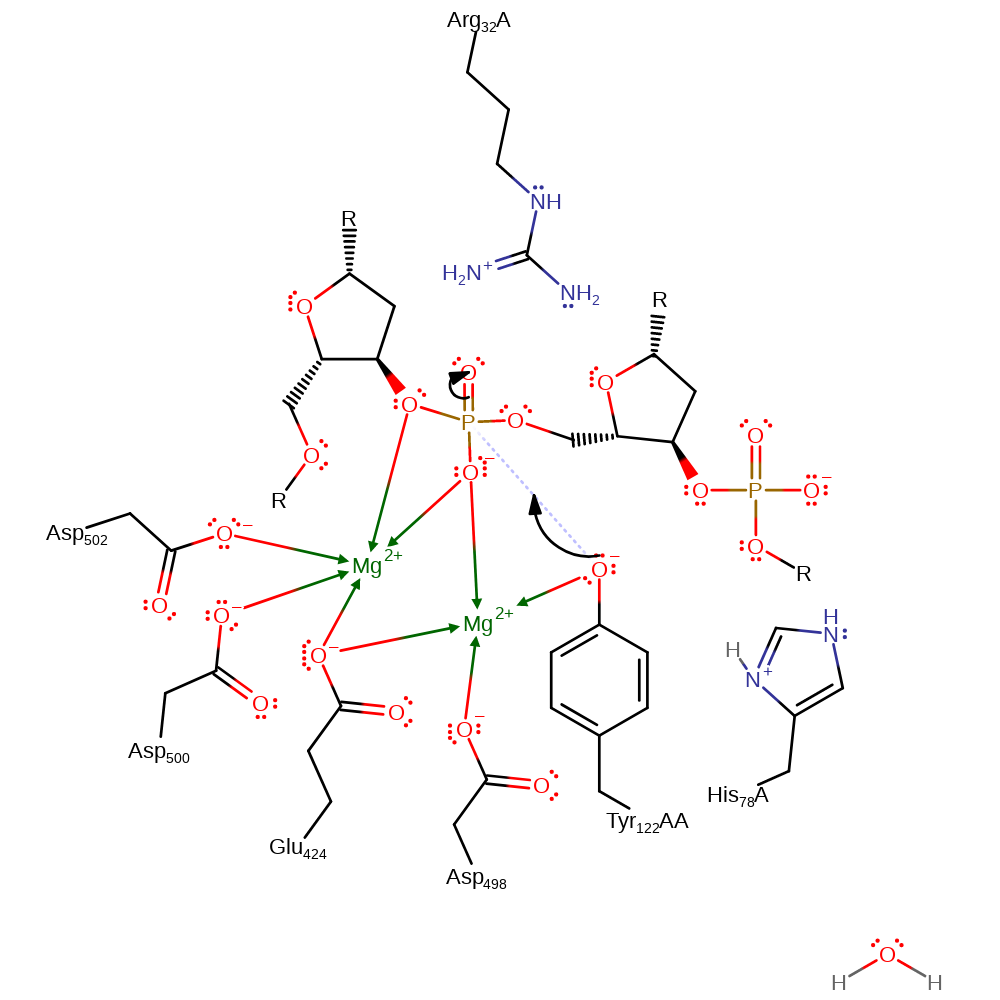
Step 2. There is nucleophilic attack from Tyr 122 on to the scissile phosphate group. This forms a penta-covalent intermediate which is stabilized by both the Mg ions and Arg 32.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu424 | metal ligand |
| Asp498 | metal ligand |
| Asp500 | metal ligand |
| Asp502 | metal ligand |
| Tyr122(93)A(AA) | metal ligand |
| Arg32(3)A | electrostatic stabiliser |
| Tyr122(93)A(AA) | nucleophile |
Chemical Components
ingold: bimolecular nucleophilic addition, intermediate formation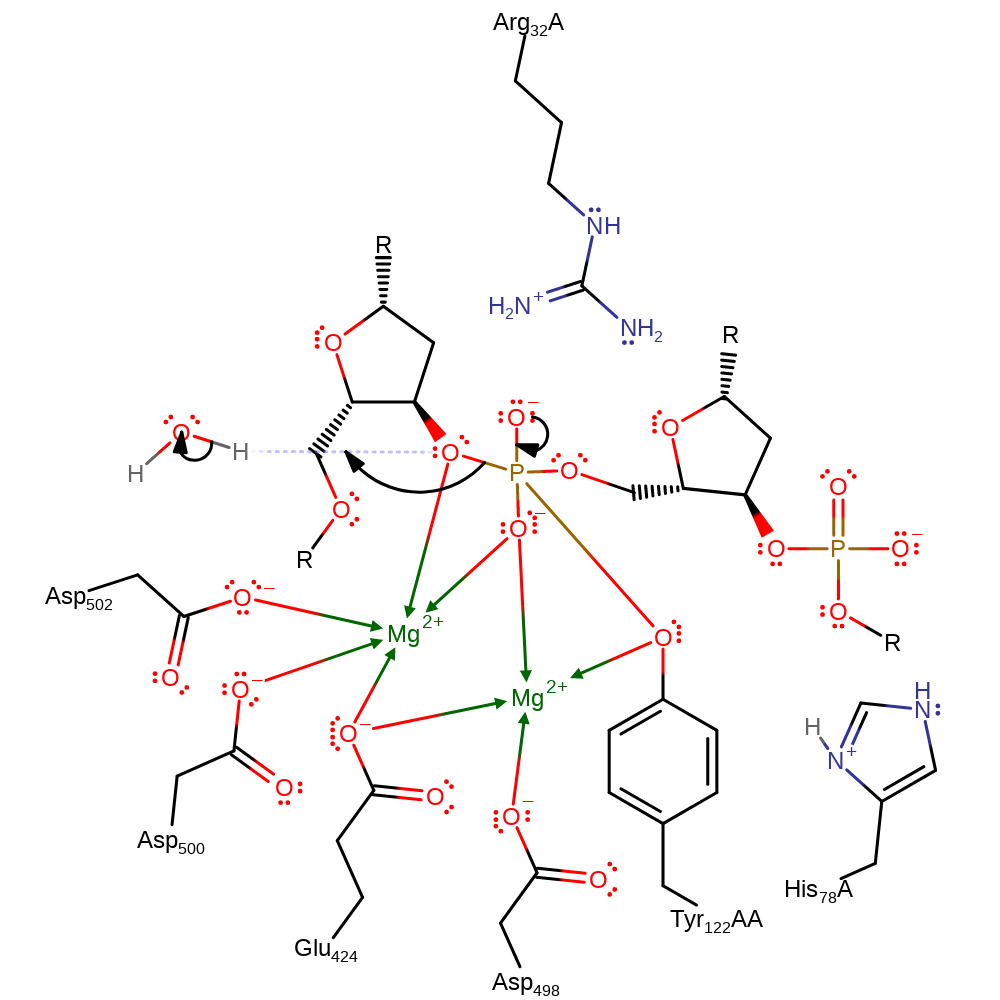
Step 3. The bond between the phosphate and the 3'hydroxyl is cleaved. The bridging oxygen is coordinated to the first Mg ion this makes the 3'-oxyanion a better leaving group. The 3'oxy-anion accepts a proton (from an unknown source). This concludes the cleavage step of the gyrase action.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu424 | metal ligand |
| Asp498 | metal ligand |
| Asp500 | metal ligand |
| Asp502 | metal ligand |
| Tyr122(93)A(AA) | metal ligand |
| Arg32(3)A | electrostatic stabiliser |
Chemical Components
ingold: intramolecular elimination, proton transfer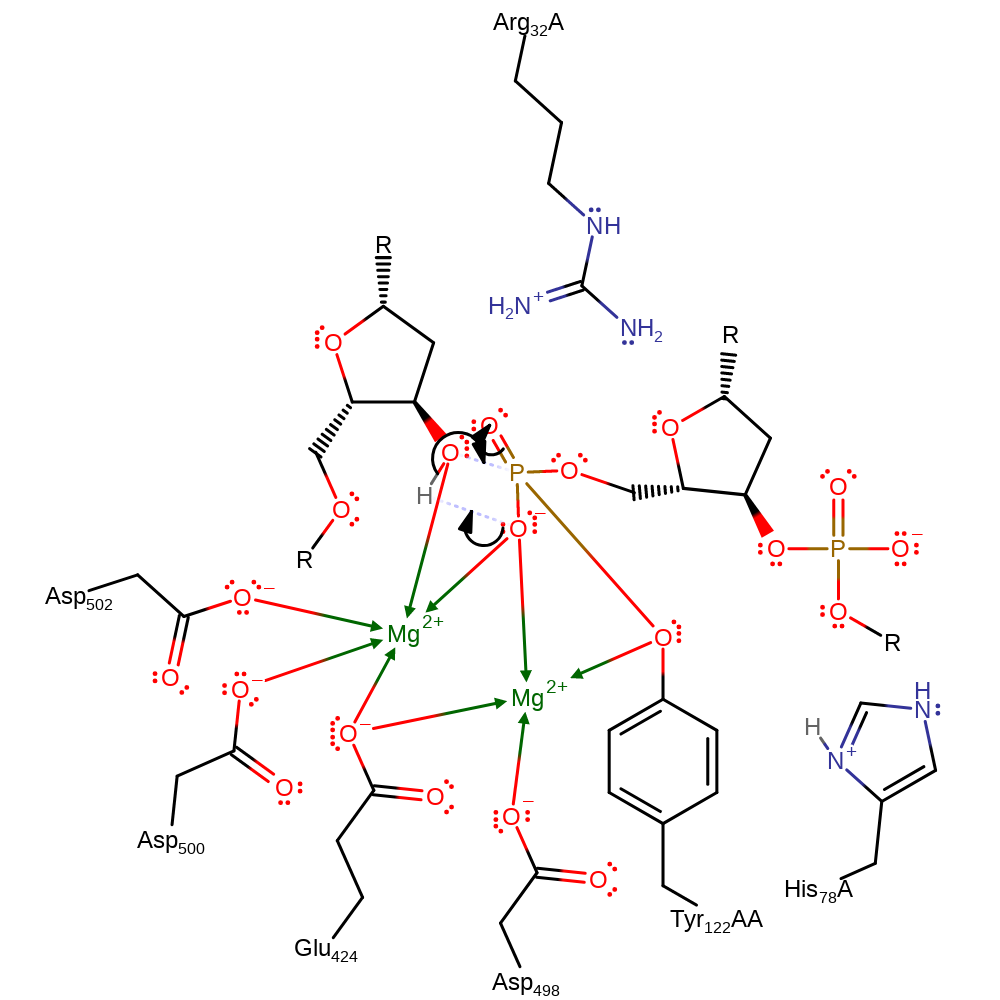
Step 4. The following steps concern the religation process, which occurs after the topological change of DNA. Intramolecular proton transfer between the phosphate and the 3' hydroxyl of the cleaved DNA promotes the nucleophilic attack of the hydroxyl on to the scissile phosphate. Forming the same intermediate as before.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu424 | metal ligand |
| Asp498 | metal ligand |
| Asp500 | metal ligand |
| Asp502 | metal ligand |
| Tyr122(93)A(AA) | metal ligand |
| Arg32(3)A | electrostatic stabiliser |
Chemical Components
proton relay, ingold: bimolecular nucleophilic additionCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu424 | metal ligand |
| Asp498 | metal ligand |
| Asp500 | metal ligand |
| Asp502 | metal ligand |
| Tyr122(93)A(AA) | metal ligand |
| Arg32(3)A | electrostatic stabiliser |
Chemical Components
proton transfer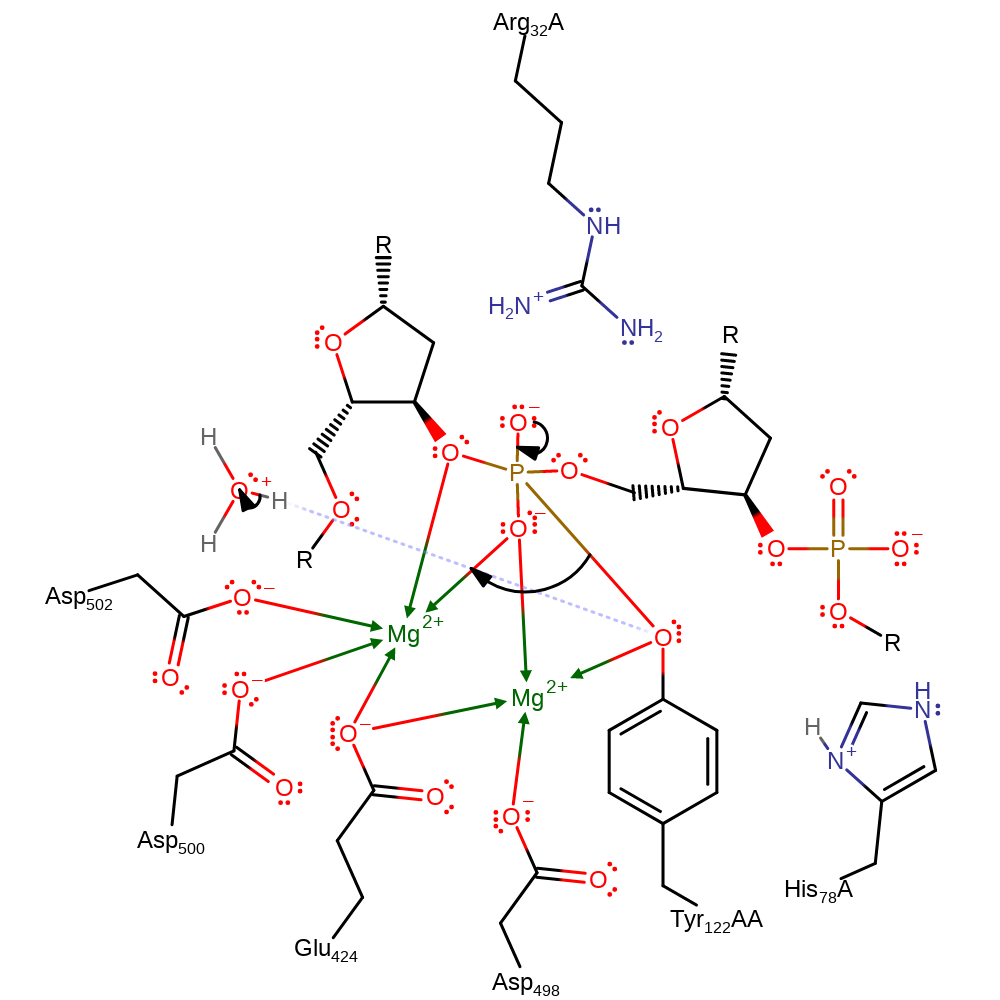
Step 6. Tyr 122 is cleaved from the phosphate and is protonated by a hydrononium ion. This completes the religation process.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu424 | metal ligand |
| Asp498 | metal ligand |
| Asp500 | metal ligand |
| Asp502 | metal ligand |
| Tyr122(93)A(AA) | metal ligand |
| Tyr122(93)A(AA) | proton acceptor, nucleofuge |






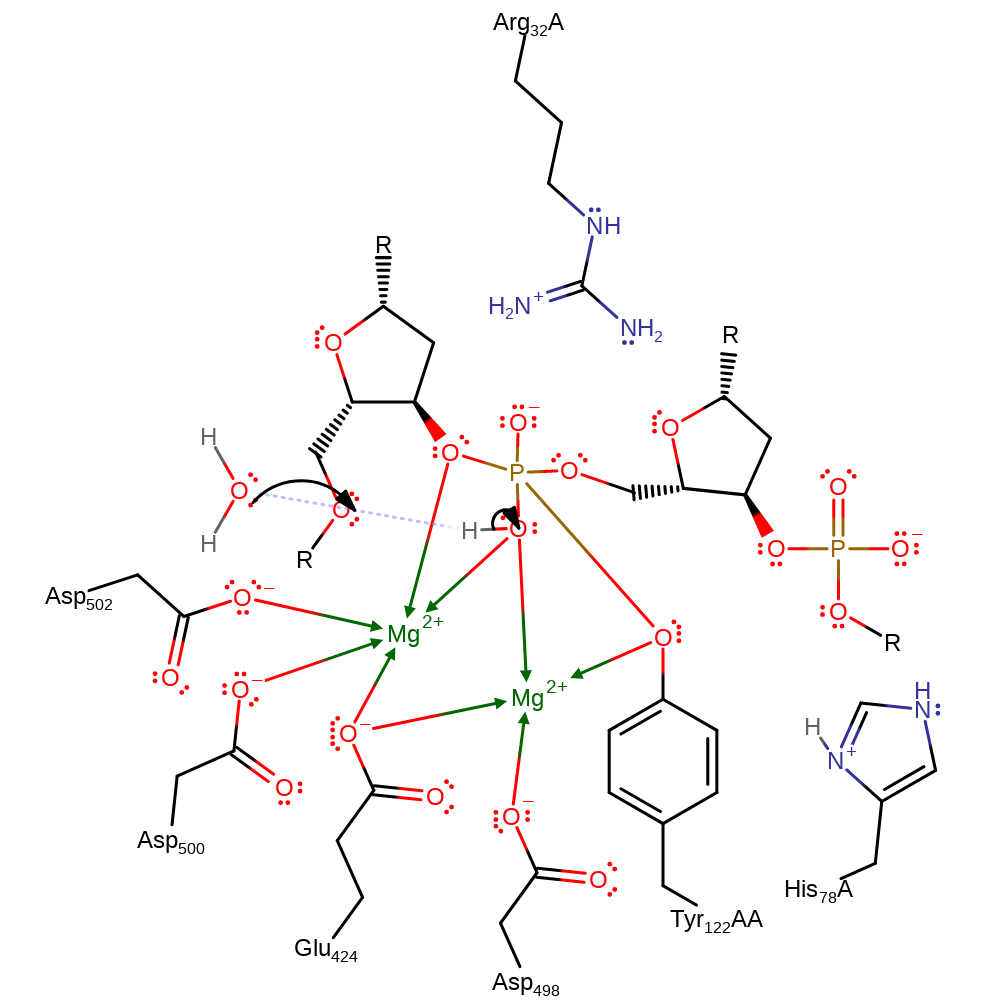
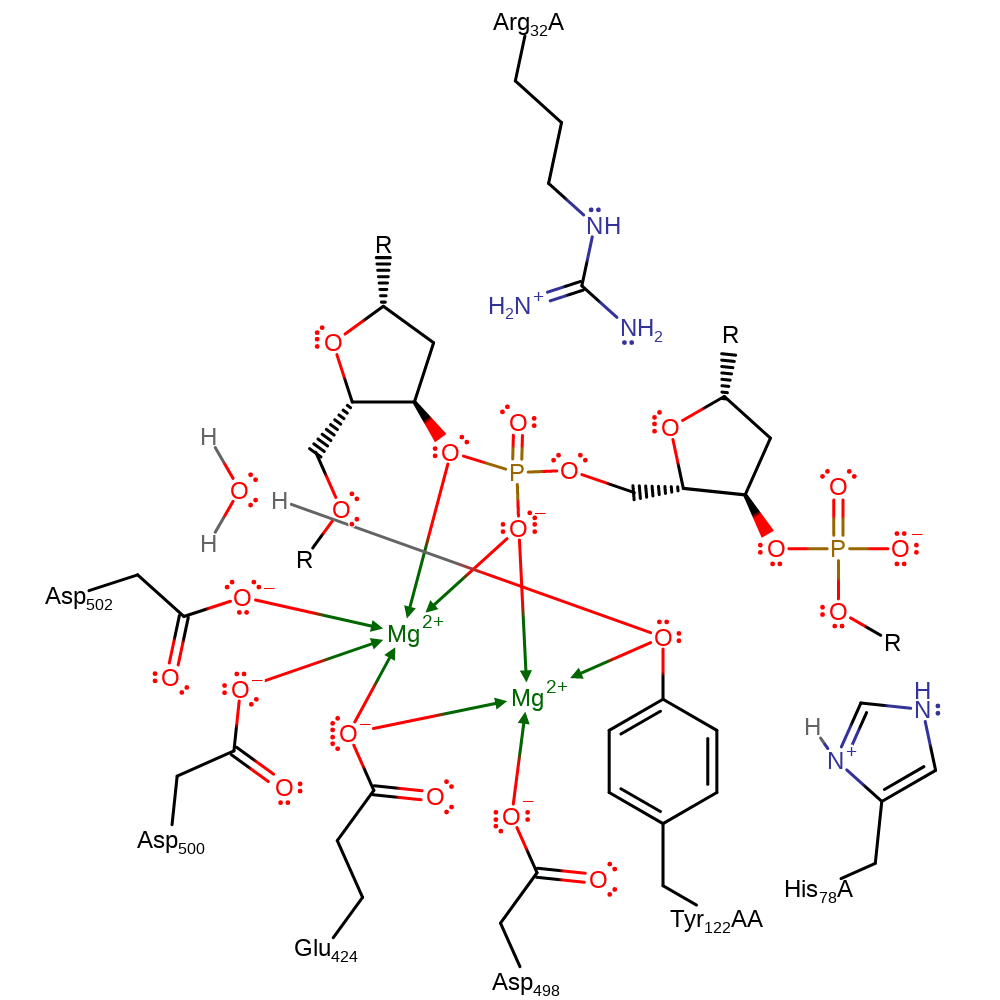 Download:
Download: