Protein disulfide oxidoreductase
DsbB isolated from Escherichia coli is able to create disulfide bonds de novo. It forms a complex with DsbA and overall there is a net formation of a disulfide bridge in DsbA and a quinone cofactor is reduced in DsbB. The quinone can either be ubiquinone (under aerobic conditions) or menaquinone (under anaerobic conditions). DsbA is a periplasmic dithiol oxidase and it is the primary disulfide bond donor in the periplasm. The disulfide bond to be produced in DsbA is between Cys30 and Cys33. Two disulfide bonds exist in DsbB in the resting state, Cys41-Cys44 and Cys104-Cys130; they are involved in the catalytic pathway.
Reference Protein and Structure
- Sequences
-
P0AEG4

P0A6M2
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Escherichia coli K-12 (Bacteria)

- PDB
-
2hi7
- Crystal structure of DsbA-DsbB-ubiquinone complex
(3.7 Å)



- Catalytic CATH Domains
-
1.20.1550.10
 (see all for 2hi7)
(see all for 2hi7)
- Cofactors
- Ubiquinone-1 (1)
Enzyme Mechanism
Introduction
DsbB contains the Cys41-Cys44 and Cys104-Cys130 disulfide bonds in the resting state. In the DsbA-DsbB complex, DsbA causes a conformational change which separates Cys104 and Cys130. The reaction begins witgh nucleophilic attack by the DsbA Cys33 thiolate on the intermolecular Cys104-Cys30 disulfide bond leading to a Cys30-Cys33 intramolecular disulfide bond and a Cys104 thiolate. The hemi-oxidised DsbB then rearranges so that the Cys41-Cys44 disulfide bond is reduced and Cys104 and Cys130 are oxidised to form a bond. The Cys44 thiolate forms a charge transfer complex with the quinone, stabilised by Arg48, and then forms an adduct through the C2 of the quinone. The negative charge is delocalised around the O2 atom and is also stabilised by Arg48. The Cys41 thiolate then acts as a nucleophile and attacks Cys44, breaking the S-C2 bond and creating a disulfide bridge and a reduced quinone molecule.
The above refers to the rapid pathway, which predominates. There is a minor pathway called the slow pathway. The two diverge after the formation of the intermolecular disulfide bond. In the slow pathway there is nucleophilic attack on the Cys41-Cys44 disulfide bond by the Cys130 thiolate, leading to a Cys41-Cys130 disulfide bond and a Cys44 thiolate. The exact mechanism by which the final disulfide bonds are generated and the quinone is reduced is unknown.
Catalytic Residues Roles
| UniProt | PDB* (2hi7) | ||
| Cys130 | SerNone(130)B | In the resting state of DsbB there is a Cys130-Cys104 disulfide bond. The Cys104–Cys130 pair is involved directly in the disulfide exchange with DsbA | electrofuge, electrophile, nucleophile |
| Cys41 | Cys41B | The Cys41-Cys44 disulfide bond is reduced in the hemi-oxidised DsbB. The Cys41 thiolate attacks the Cys44-quinone adduct to reform the Cys41-Cys44 disulfide bond. | electrofuge, electrophile, nucleophile, nucleofuge |
| Cys44 | Cys44B | The Cys41-Cys44 disulfide bond is reduced in the hemi-oxidised DsbB. The Cys44 thiolate forms a charge transfer complex with the quinone and then an adduct. The sulfur acts as the electrophile in the nucleophilic substitution by Cys41, thus reforming the disulfide bond. | electrophile, electrofuge, nucleofuge, nucleophile |
| Arg48 | Arg48B | Arg48 stabilises the charge transfer complex between the Cys44 thiolate and the quinone. It also stabilises the adduct formed between the two. | electrostatic stabiliser |
| Cys104 | Cys104B | The Cys104-Cys130 bond is cleaved by nucleophilic attack by the Cys30 thiolate, forming a Cys104-Cys30 bond. This bond is then cleaved after nucleophilic attack by the Cys33 thiolate, leaving Cys104 as a thiolate. The rearrangement of the hemi-oxidised DsbB allows the Cys104-Cys130 bond to reform when the Cys41-Cys44 bond is reduced. | covalently attached, nucleofuge, nucleophile |
Chemical Components
bimolecular nucleophilic substitution, overall product formed, bimolecular nucleophilic addition, michael addition, overall reactant used, intermediate formation, enzyme-substrate complex formation, proton transfer, native state of enzyme regenerated, enzyme-substrate complex cleavageReferences
- Inaba K et al. (2006), Cell, 127, 789-801. Crystal Structure of the DsbB-DsbA Complex Reveals a Mechanism of Disulfide Bond Generation. DOI:10.1016/j.cell.2006.10.034. PMID:17110337.
- Inaba K et al. (2009), EMBO J, 28, 779-791. Dynamic nature of disulphide bond formation catalysts revealed by crystal structures of DsbB. DOI:10.1038/emboj.2009.21. PMID:19214188.
- Inaba K et al. (2008), Biochim Biophys Acta, 1783, 520-529. Structure and mechanisms of the DsbB–DsbA disulfide bond generation machine. DOI:10.1016/j.bbamcr.2007.11.006. PMID:18082634.
- Inaba K et al. (2006), Proc Natl Acad Sci U S A, 103, 287-292. Critical role of a thiolate-quinone charge transfer complex and its adduct form in de novo disulfide bond generation by DsbB. DOI:10.1073/pnas.0507570103. PMID:16384917.
- Kishigami S et al. (1996), Genes Cells, 1, 201-208. Roles of cysteine residues of DsbB in its activity to reoxidize DsbA, the protein disulphide bond catalyst ofEscherichia coli. DOI:10.1046/j.1365-2443.1996.d01-233.x. PMID:9140064.

Step 1. DsbB contains the Cys41-Cys44 and Cys104-Cys130 disulfide bonds in the resting state. In the DsbA-DsbB complex, DsbA causes a conformational change which separates Cys104 and Cys130. The rapid pathway begins when Cys33 of DsbA attacks the intermolecular Cys104 (DsbB) - Cys30 (DbsA) disulfide bond. This forms the Cys30 - Cys33 disulfide product in DsbA.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Cys104B | covalently attached |
| Cys104B | nucleofuge |
Chemical Components
ingold: bimolecular nucleophilic substitution, overall product formed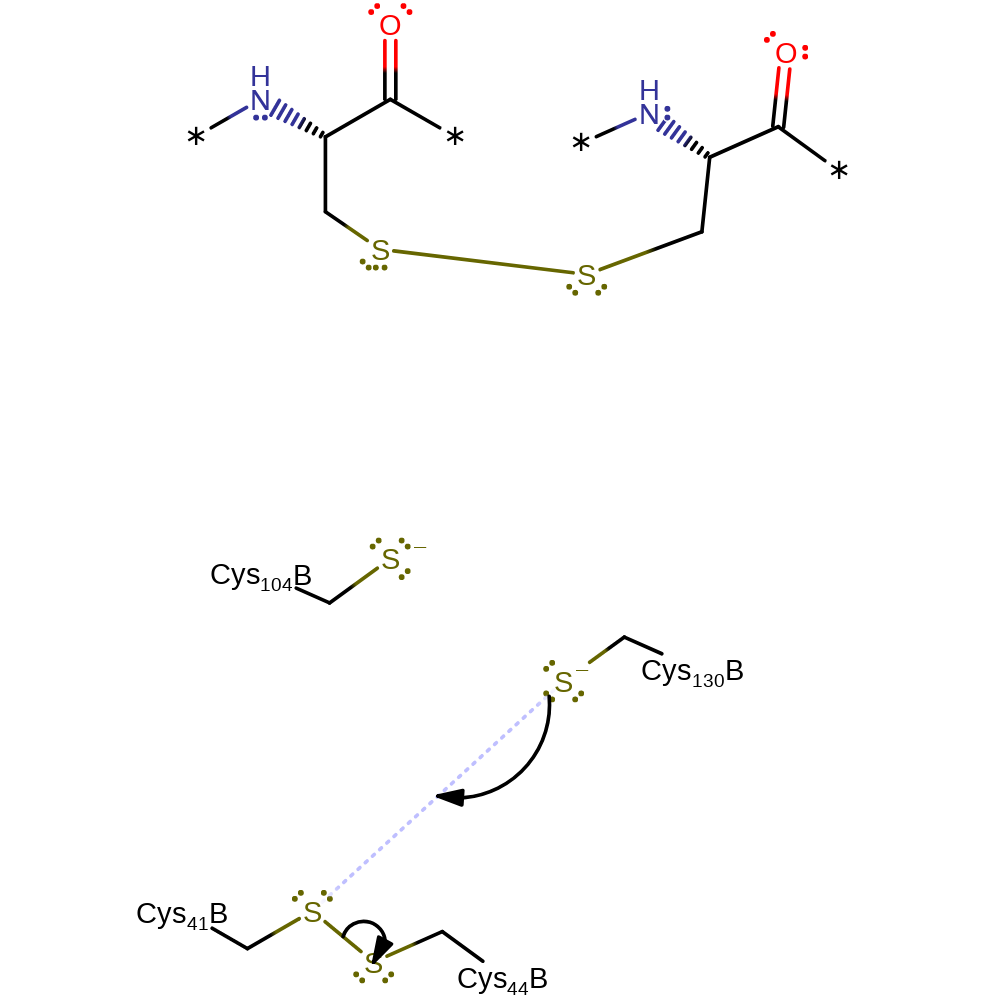
Step 2. Cys130 then initiates nucleophilic attack the Cys44-Cys41 disulfide bond. This is the first step in a dithiol disulfide exchange between the active site residues.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Cys41B | electrofuge |
| SerNone(130)B | nucleophile |
| Cys41B | electrophile |
| Cys44B | nucleofuge |
Chemical Components
ingold: bimolecular nucleophilic substitution
Step 3. There is nucleophilic attack of Cys104 on the Cys130-Cys41 disulfide bond. In comparison to the original resting state of DsbB, the Cys41-Cys44 disulfide bond has been reduced by two electrons.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| SerNone(130)B | electrofuge, electrophile |
| Cys41B | nucleofuge |
| Cys104B | nucleophile |
Chemical Components
ingold: bimolecular nucleophilic substitution
Step 4. Cys44 forms a charge transfer complex with ubiquinone before initiating nucleophilic attack and forming an adduct. Arg48 stabilises Cys44 and the adduct formed.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg48B | electrostatic stabiliser |
| Cys44B | nucleophile |
Chemical Components
ingold: bimolecular nucleophilic addition, michael addition, overall reactant used, intermediate formation, enzyme-substrate complex formation, proton transfer
Step 5. The S-C covalent linkage in the adduct is attacked by Cys41, reforming the Cys41-Cys44 disulfide bond and resulting in a quinol. The quinol is subsequently protonated to complete the reaction.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg48B | electrostatic stabiliser |
| Cys44B | electrophile, electrofuge |
| Cys41B | nucleophile |




 Download:
Download: