Esterase
Esterase (EST), isolated from Pseudomonas putida, catalyses the hydrolysis of methyl esters. Of particular interest is the stereoselective hydrolysis of DL-beta-acetylthioisobutyrate acid (DL-MATI) to D-beta-acetylthioisobutyric acid (DAT). This is because DAT is an important intermediate in the synthesis of the medicinally important angiotensin-converting enzyme inhibitors. The mechanism of action of EST is similar to the serine protease mechanism.
Reference Protein and Structure
- Sequence
-
Q3HWU8

 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Pseudomonas putida (Bacteria)

- PDB
-
1zoi
- Crystal Structure of a Stereoselective Esterase from Pseudomonas putida IFO12996
(1.6 Å)



- Catalytic CATH Domains
-
3.40.50.1820
 (see all for 1zoi)
(see all for 1zoi)
Enzyme Mechanism
Introduction
His256 deprotonates Ser97 and is stabilised by Asp227. Ser97 is then involved in nucleophilic attack on the carbonyl carbon of D-MALTI. The resulting tetrahedral oxyanion is stabilised by hydrogen bonds to the backbone amines of Trp31 and Thr98, which together form the oxyanion hole. The intermediate collapses to eliminate methoxide, which is protonated by His256. His256 then deprotonates water and is again stabilised by Asp227. The resulting hydroxide is involved in nucleophilic attack on the carbonyl carbon of the acylenzyme intermediate. The tetrahedral intermediate is stabilised by the oxyanion hole and then collapses to eliminate Ser97, which is protonated by His256, and to form DAT.
Catalytic Residues Roles
| UniProt | PDB* (1zoi) | ||
| His256 | His256A | His256 acts as a general acid and a general base during the reaction. It deprotonates the nucleophiles (Ser97 and water) and protonates the leaving groups (methoxide and deprotonated Ser97). | proton acceptor, proton donor |
| Thr98 (main-N) | Thr98A (main-N) | Thr98 forms part of the oxyanion hole and stabilises the tetrahedral intermediate by hydrogen bonding to the alkoxide. | electrostatic stabiliser |
| Ser97 | Ser97A | Ser97 is deprotonated by His256 and then acts as the nucleophile for attack on the carbonyl of the ester. It is the leaving group in the deacylation reaction and is protonated by His256. | nucleofuge, nucleophile, proton acceptor, proton donor |
| Ala122 (main-C) | Ala122A (main-C) | The backbone carbonyl of Ala122 forms a hydrogen bond to the epsilon-1 C-H of His256. This weakens the epsilon-2 N-H bond in the protonated histidine, assisting general acid catalysis. | electrostatic stabiliser |
| Trp31 (main-N) | Trp31A (main-N) | Trp31 forms part of the oxyanion hole and stabilises the tetrahedral intermediate by hydrogen bonding to the alkoxide. | electrostatic stabiliser |
| Asp227 | Asp227A | Asp227 forms a hydrogen bond to the delta-1 N-H of His256. This stabilises the protonated state of His256. | electrostatic stabiliser |
Chemical Components
proton transfer, bimolecular nucleophilic addition, intermediate formation, enzyme-substrate complex formation, overall reactant used, unimolecular elimination by the conjugate base, intermediate collapse, overall product formed, enzyme-substrate complex cleavage, native state of enzyme regeneratedReferences
- Elmi F et al. (2005), J Bacteriol, 187, 8470-8476. Stereoselective Esterase from Pseudomonas putida IFO12996 Reveals / Hydrolase Folds for D- -Acetylthioisobutyric Acid Synthesis. DOI:10.1128/jb.187.24.8470-8476.2005. PMID:16321951.
- Hedstrom L (2002), Chem Rev, 34, 4501-4524. Serine Protease Mechanism and Specificity. DOI:10.1002/chin.200306269. PMID:12475199.
- Heikinheimo P et al. (1999), Structure, 7, R141-R146. Of barn owls and bankers: a lush variety of α/β hydrolases. DOI:10.1016/s0969-2126(99)80079-3. PMID:10404588.
- Derewenda ZS et al. (1994), J Mol Biol, 241, 83-93. (His)Cε-H···O=C< Hydrogen Bond in the Active Sites of Serine Hydrolases. DOI:10.1006/jmbi.1994.1475. PMID:8051710.

Step 1. His256 deprotonates Ser97 which activates it to nucleophilically attack the carbonyl carbon.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Thr98A (main-N) | electrostatic stabiliser |
| Trp31A (main-N) | electrostatic stabiliser |
| Ala122A (main-C) | electrostatic stabiliser |
| Asp227A | electrostatic stabiliser |
| Ser97A | nucleophile, proton donor |
| His256A | proton acceptor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, intermediate formation, enzyme-substrate complex formation, overall reactant used
Step 2. The intermediate collapses to eliminate methoxide, which is protonated by His256.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Trp31A (main-N) | electrostatic stabiliser |
| Thr98A (main-N) | electrostatic stabiliser |
| Ala122A (main-C) | electrostatic stabiliser |
| Asp227A | electrostatic stabiliser |
| His256A | proton donor |
Chemical Components
ingold: unimolecular elimination by the conjugate base, proton transfer, intermediate collapse, overall product formed
Step 3. His256 abstracts a proton from a water which then attacks the carbonyl carbon of the acylenzyme intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Trp31A (main-N) | electrostatic stabiliser |
| Thr98A (main-N) | electrostatic stabiliser |
| Ala122A (main-C) | electrostatic stabiliser |
| Asp227A | electrostatic stabiliser |
| His256A | proton acceptor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, intermediate formation, overall reactant used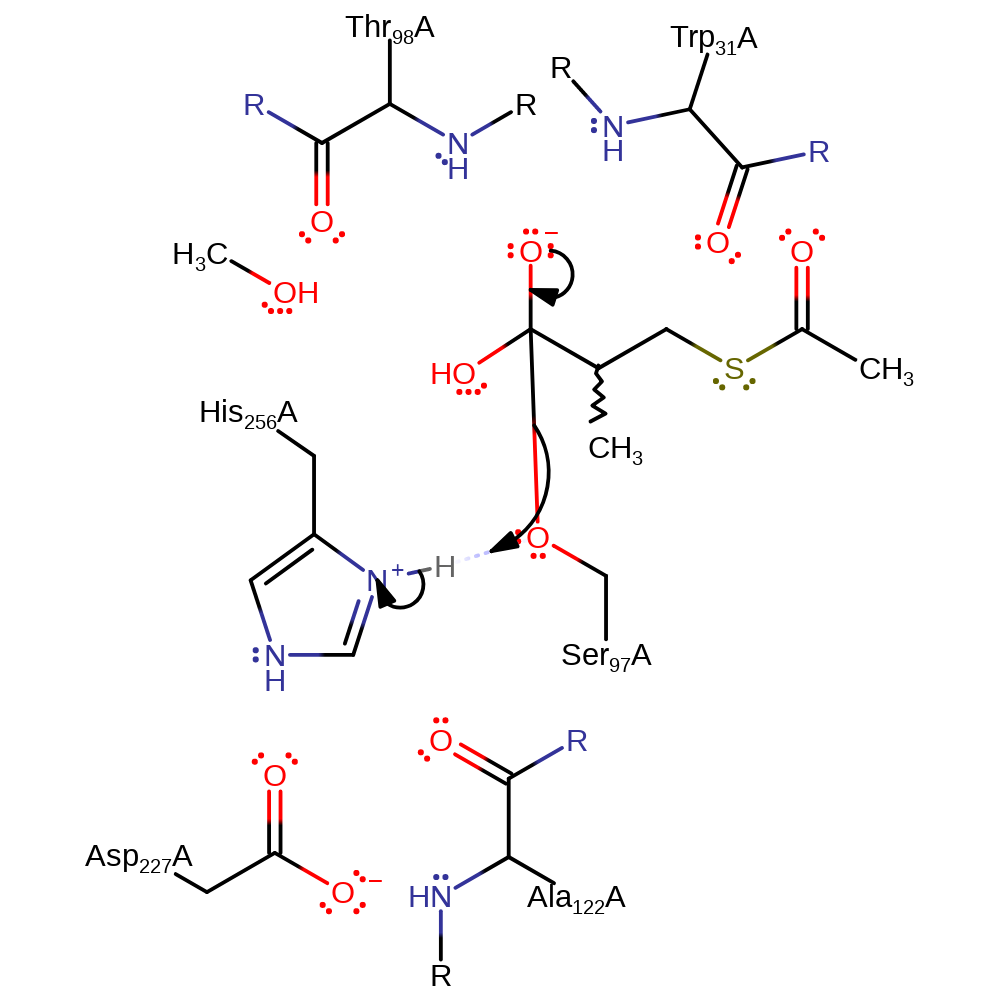
Step 4. The tetrahedral intermediate is stabilised by the oxyanion hole and then collapses to eliminate Ser97, which is protonated by His256.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Trp31A (main-N) | electrostatic stabiliser |
| Thr98A (main-N) | electrostatic stabiliser |
| Ala122A (main-C) | electrostatic stabiliser |
| Asp227A | electrostatic stabiliser |
| Ser97A | nucleofuge, proton acceptor |
| His256A | proton donor |




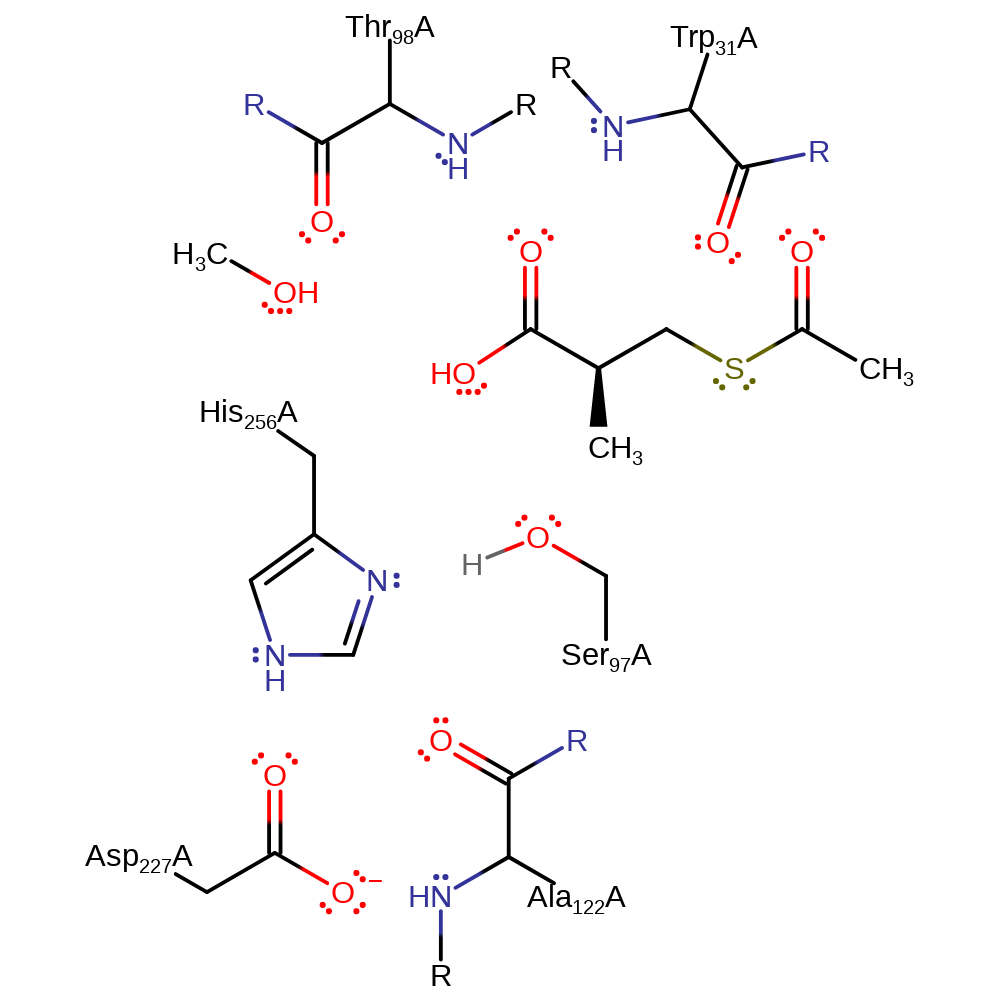 Download:
Download: