Repressor LexA
LexA repressor (LexA) from Escherichia coli hydrolyses the specific peptide bond between Alanine and Glycine residues. It is of the peptidase S24 family. It can take two conformations; cleavable and non cleavable. In the non cleavable conformer, the Ala 84 - Gly 85 peptide bond lies about 20 angstroms away from the active site. In the cleavable conformer, part of the protein, the Ala 84 - Gly 85 peptide bond lies directly adjacent to the active site. In this case, LexA will cleave itself and become denatured. This function allows LexA to control the SOS response of E. coli to conditions that damage DNA or inhibit DNA replication.
Reference Protein and Structure
- Sequence
-
P0A7C2
 (3.4.21.88)
(3.4.21.88)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Escherichia coli K-12 (Bacteria)

- PDB
-
1jhf
- LEXA G85D MUTANT
(1.8 Å)



- Catalytic CATH Domains
-
2.10.109.10
 (see all for 1jhf)
(see all for 1jhf)
Enzyme Mechanism
Introduction
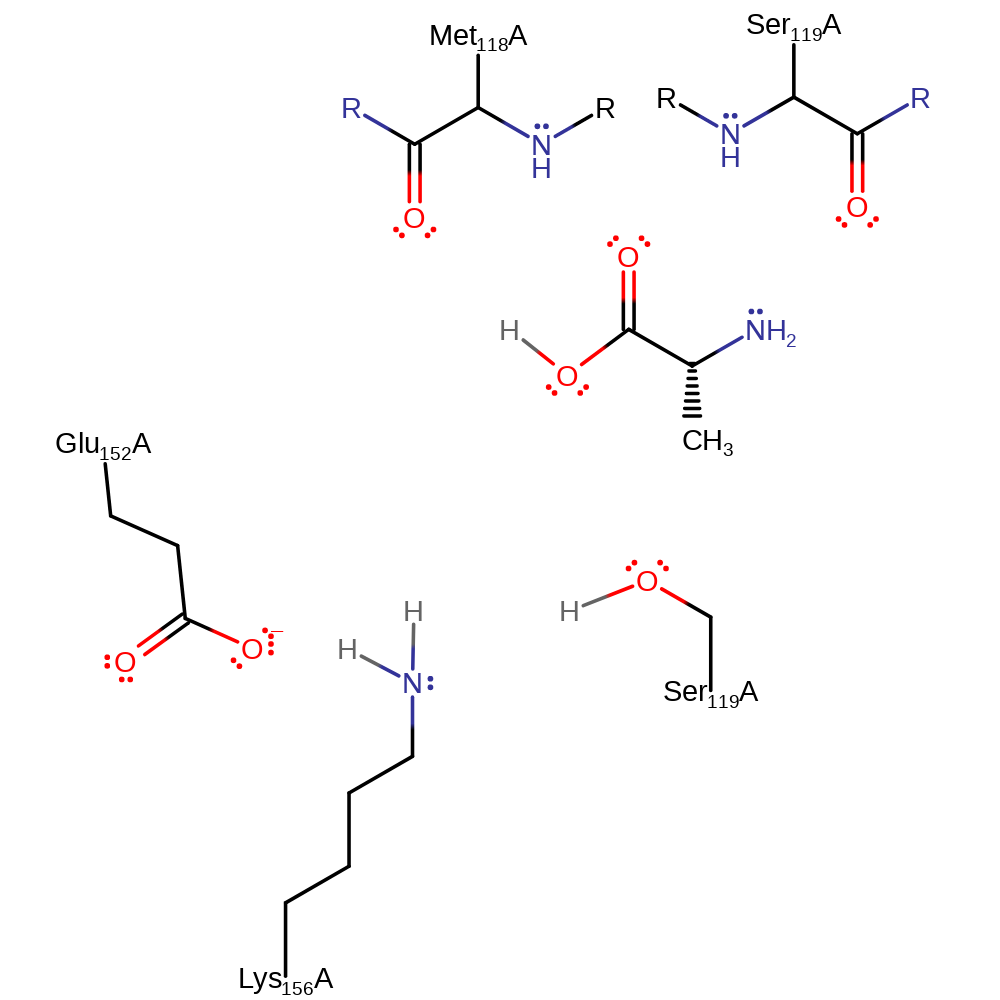
Ser 119 is activated on being deprotonated by Lys 156, and nucleophilically attacks the carbonyl of the scissile peptide bond, forming a negatively charged tetrahedral intermediate. The protonated form of Lys 156 is stabilised through hydrogen bonding to the side chain of Glu 152. The intermediate is stabilised by the oxyanion hole formed by the backbone NH's of Ser 119 and Met 118. The oxyanion hole itself is stabilised by hydrogen bonding to the main chain of Gly 117, the side chain of Asp 127 and a solvent molecule. The carbonyl reforms with the loss of an amino group, which is protonated by the previously protonated Lys 156. Lys 156 then activates a water molecule, acting as a general base, and the water molecule nucleophilically attacks the carbonyl of the scissile bond, forming a negatively charged, tetrahedral intermediate. This intermediate is again stabilised by the oxyanion hole. The carbonyl is reformed again to form the product, and the leaving group, Ser 119, is protonated by the protonated Lys 156.
Catalytic Residues Roles
| UniProt | PDB* (1jhf) | ||
| Ser119 | Ser119A | Ser 119 is activated by Lys 156 and nucleophilically attacks the carbonyl of the scissile peptide bond, and forms a tetrahedral intermediate. The main chain amide N atom of Ser 119 makes up part of the oxyanion hole, and stabilises the tetrahedral intermediate. | nucleofuge, nucleophile, proton acceptor, proton donor |
| Glu152 | Glu152A | The gamma carboxylate of Glu 152 stabilises the protonated Lys 156 residue. | electrostatic stabiliser |
| Met118 (main-N) | Met118A (main-N) | Met 118 (along with Ser 119) forms the oxyanion hole and stabilises the tetrahedral intermediate. | electrostatic stabiliser |
| Lys156 | Lys156A | Lys 156 activates Ser 119 by deprotonating it. It then donates the proton to the leaving group. Lys 156 then deprotonates a water molecule, and on expulsion of Ser 119 from the tetrahedral intermediate, re-protonates Ser 119. | proton acceptor, proton donor |
Chemical Components
proton transfer, bimolecular nucleophilic addition, intermediate formation, overall reactant used, rate-determining step, unimolecular elimination by the conjugate base, intermediate collapse, overall product formed, native state of enzyme regeneratedReferences
- Luo Y et al. (2001), Cell, 106, 585-594. Crystal structure of LexA: a conformational switch for regulation of self-cleavage. PMID:11551506.
- Slilaty SN et al. (1987), Proc Natl Acad Sci U S A, 84, 3987-3991. Lysine-156 and serine-119 are required for LexA repressor cleavage: a possible mechanism. DOI:10.1073/pnas.84.12.3987. PMID:3108885.
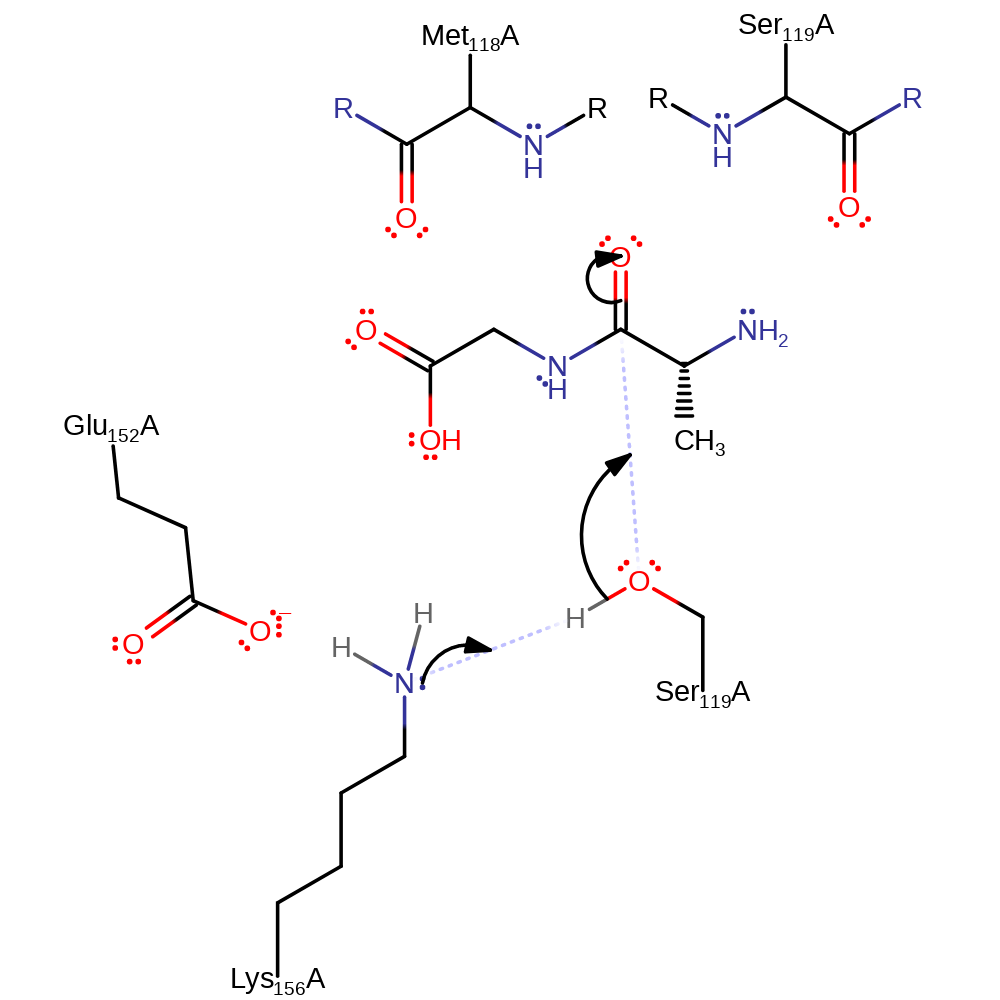
Step 1. Ser 119 is activated when it is deprotonated by Lys 156, and nucleophilically attacks the carbonyl of the scissile peptide bond, forming a negatively charged tetrahedral intermediate. Glu 152 hydrogen bonds to Lys 156 to stabilise it and enable it to accept a proton.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Met118A (main-N) | electrostatic stabiliser |
| Glu152A | electrostatic stabiliser |
| Ser119A (main-N) | electrostatic stabiliser |
| Ser119A | proton donor |
| Lys156A | proton acceptor |
| Ser119A | nucleophile |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, intermediate formation, overall reactant used, rate-determining step
Step 2. The oxyanion initiates an elimination which results in the cleavage of the peptide bond. The N-terminal product is then protonated by Lys 156.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Met118A (main-N) | electrostatic stabiliser |
| Ser119A (main-N) | electrostatic stabiliser |
| Glu152A | electrostatic stabiliser |
| Lys156A | proton donor |
Chemical Components
ingold: unimolecular elimination by the conjugate base, proton transfer, intermediate collapse, overall product formed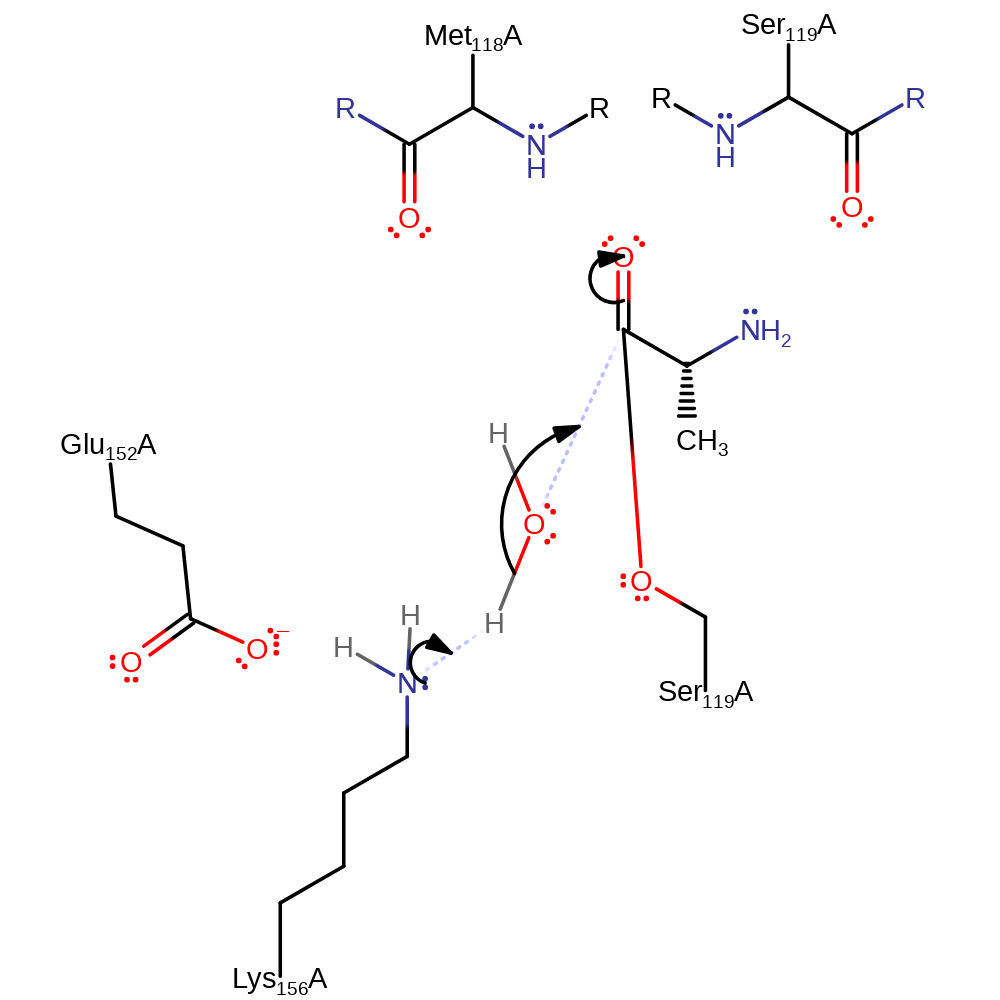
Step 3. Lys 156 abstracts a proton from a water, activating it to attack the carbon of the ester bond in a nucleophilic addition.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Met118A (main-N) | electrostatic stabiliser |
| Ser119A (main-N) | electrostatic stabiliser |
| Glu152A | electrostatic stabiliser |
| Lys156A | proton acceptor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, intermediate formation, overall reactant used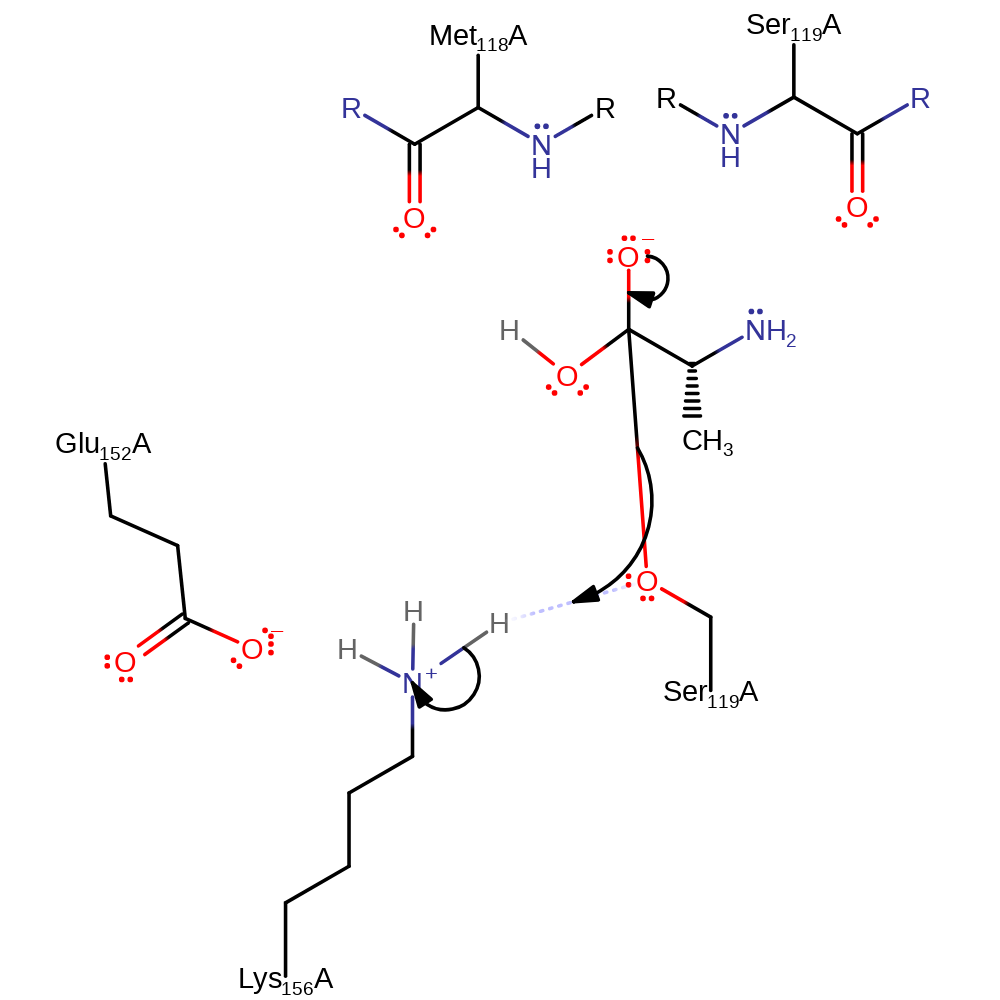
Step 4. The tetrahedral intermediate collapse again resulting the release of Ser 119 which then accepts a proton from Lys 156 which returns the enzyme to its native state.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Met118A (main-N) | electrostatic stabiliser |
| Ser119A (main-N) | electrostatic stabiliser |
| Glu152A | electrostatic stabiliser |
| Ser119A | nucleofuge, proton acceptor |
| Lys156A | proton donor |



 Download:
Download: