Creatininase
Creatininase (creatinine amidohydrolase) from Pseudomonas putida catalyses the conversion of creatinine to creatine. It is a member of the urease-related amidohydrolase superfamily. The enzyme enables the bacteria to use creatinine as a source of carbon and nitrogen. The biological molecule is trimer of dimers. The active site of each monomer contains a binuclear metal centre located at the bottom of a cleft. In the native form of the enzyme both metal sites are occupied by Zn2+ ions.
Reference Protein and Structure
- Sequence
-
P83772
 (3.5.2.10)
(3.5.2.10)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Pseudomonas putida (Bacteria)

- PDB
-
1j2u
- Creatininase Zn
(1.85 Å)



- Catalytic CATH Domains
-
3.40.50.10310
 (see all for 1j2u)
(see all for 1j2u)
- Cofactors
- Zinc(2+) (2)
Enzyme Reaction (EC:3.5.2.10)
Enzyme Mechanism
Introduction
A water molecule bridging the two metal ions is likely to form a hydroxide ion which acts as the nucleophile. The hydroxide attacks the carbonyl carbon of the creatinine substrate and is then deprotonated by His 178. The tetrahedral intermediate is stabilised by coordination to the two metal ions. Ring opening occurs simultaneously with the protonation of the amide nitrogen on the intermediate by His 178.
Catalytic Residues Roles
| UniProt | PDB* (1j2u) | ||
| Glu34, His120, Asp45, His36, Glu183 | Glu34A, His120A, Asp45A, His36A, Glu183A | Forms zinc binding site. | metal ligand |
| Glu122 | Glu122A | Glu122 forms a hydrogen bond to the water molecule part of the zinc coordination sphere and likely protonates creatine to make the kinetically favourable product. | electrostatic stabiliser |
| His178 | His178A | Deprotonates the hydroxide that is part of the oxyanion intermediate and then protonates the amide group to initiate an elimination. | proton acceptor, proton donor |
Chemical Components
bimolecular nucleophilic addition, intermediate formation, overall reactant used, proton transfer, unimolecular elimination by the conjugate base, intermediate collapse, overall product formedReferences
- Yoshimoto T et al. (2004), J Mol Biol, 337, 399-416. Crystal Structures of Creatininase Reveal the Substrate Binding Site and Provide an Insight into the Catalytic Mechanism. DOI:10.1016/j.jmb.2004.01.022. PMID:15003455.
- Jitonnom J et al. (2017), Biochemistry, 56, 6377-6388. Quantum Mechanics/Molecular Mechanics Simulations Identify the Ring-Opening Mechanism of Creatininase. DOI:10.1021/acs.biochem.7b01032. PMID:29140090.

Step 1. The zinc-bound water performs a nucleophilic attack attack on the carbon of the carbonyl bond.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu122A | electrostatic stabiliser |
| Glu34A | metal ligand |
| His36A | metal ligand |
| Asp45A | metal ligand |
| His120A | metal ligand |
| Glu183A | metal ligand |
Chemical Components
ingold: bimolecular nucleophilic addition, intermediate formation, overall reactant usedCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu34A | metal ligand |
| His36A | metal ligand |
| Asp45A | metal ligand |
| His120A | metal ligand |
| Glu183A | metal ligand |
| Glu122A | electrostatic stabiliser |
| His178A | proton acceptor |
Chemical Components
proton transfer, intermediate formation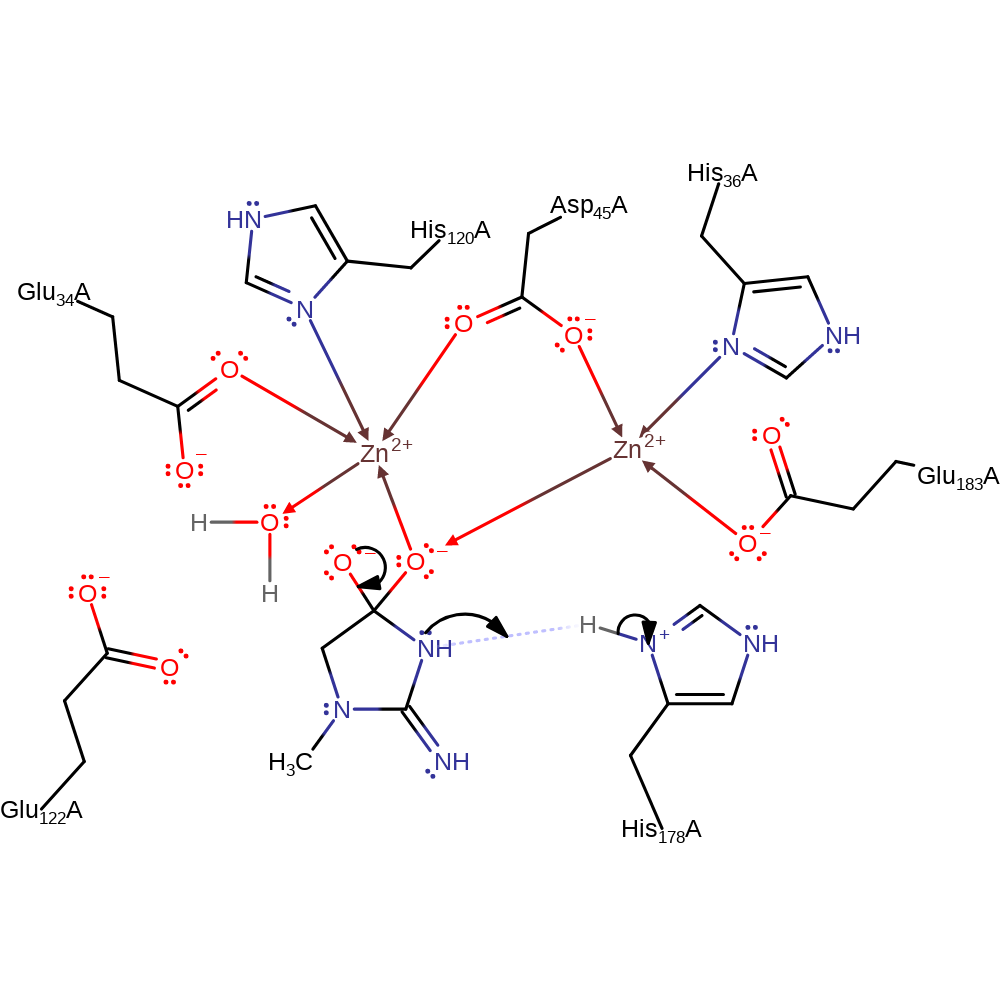
Step 3. The oxyanion initiates an elimination which results in the cleavage of the amide bond and the nitrogen accepts a proton from His178.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu34A | metal ligand |
| His36A | metal ligand |
| Asp45A | metal ligand |
| His120A | metal ligand |
| Glu183A | metal ligand |
| Glu122A | electrostatic stabiliser |
| His178A | proton donor |




 Download:
Download: