L-fuculose-phosphate aldolase
L-fuculose-phosphate aldolase (FucA) catalyses the cleavage of L-fuculose-1-phosphate to dihydroxyacetone phosphate (DHAP) and L-lactaldehyde , a conversion from a six carbon unit to a three carbon unit. FucA is a homotetramer with 215 amino acid residues and one zinc ion per subunit.
The enzymatic activity is dependent upon on the presence of a metal ion, a characteristic of the enzyme class II aldolases, to which FucA belongs. These aldolases are of great interest to synthetic chemists for the their ability to catalyse the stereoselective synthesis of sugars in a much cleaner reaction than that possible by classical organic synthesis.
Reference Protein and Structure
- Sequence
-
P0AB87
 (4.1.2.17)
(4.1.2.17)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Escherichia coli K-12 (Bacteria)

- PDB
-
2fua
- L-FUCULOSE 1-PHOSPHATE ALDOLASE CRYSTAL FORM T WITH COBALT
(2.0 Å)



- Catalytic CATH Domains
-
3.40.225.10
 (see all for 2fua)
(see all for 2fua)
- Cofactors
- Zinc(2+) (1) Metal MACiE
Enzyme Reaction (EC:4.1.2.17)
Enzyme Mechanism
Introduction
In the non substrate bound enzyme, Glu73, the proposed catalytic base is deprotonated and coordinated to the Zn centre. When dihydroxyacetone or 1-fructose binds, in what is assumed an open configuration, the residue is pushed aside into a non polar environment. This increases its pKa such that the residue can readily deprotonated either the C3 atom of dihydroxyacetone or O4 atom of 1-fructose of the reverse or forward reaction. Glu73 then 'delivers' the proton to the other side of the formed or broken bond, respectively.
Catalytic Residues Roles
| UniProt | PDB* (2fua) | ||
| Glu73 | Glu73A | The residue acts as both the catalytic acid and base within the reversible reaction, first deprotonating the O4 atom of 1-fructose in the forward direction and then donating a proton to the anionic dihydroxyacetone. In the reverse direction, Glu73 abstracts a proton from the C2 position of dihydroxyacetone, which then attacks the carbonyl of L-lactaldehyde in an aldol addition. Glu73 then protonates the newly formed hydroxyl. In the non-substrate bound form of the protein, it forms part of the zinc binding site. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
| His155, His94, His92 | His155A, His94A, His92A | Forms part of the zinc binding site. | metal ligand |
| Tyr209, Tyr113 | Tyr209A(AB), Tyr113A(AB) | Functions as independent dipoles which assist in carbonyl binding and in stabilising transition states along the aldol reaction coordinate. | electrostatic stabiliser, transition state stabiliser |
Chemical Components
bimolecular elimination, overall reactant used, overall product formed, intermediate formation, proton transfer, native state of enzyme regenerated, intermediate terminatedReferences
- Joerger AC et al. (2000), Biochemistry, 39, 6033-6041. Catalytic Action of Fuculose 1-Phosphate Aldolase (Class II) As Derived from Structure-Directed Mutagenesis†,‡. DOI:10.1021/bi9927686. PMID:10821675.
- Karthik L et al. (2013), J Struct Funct Genomics, 14, 59-70. Crystal structure analysis of l-fuculose-1-phosphate aldolase from Thermus thermophilus HB8 and its catalytic action: as explained through in silico. DOI:10.1007/s10969-013-9156-8. PMID:23744484.
- Garrabou X et al. (2010), Chemistry, 16, 10691-10706. Structure-Guided Minimalist Redesign of the L-Fuculose-1-Phosphate Aldolase Active Site: Expedient Synthesis of Novel Polyhydroxylated Pyrrolizidines and their Inhibitory Properties Against Glycosidases and Intestinal Disaccharidases. DOI:10.1002/chem.201000714. PMID:20661960.
- Joerger AC et al. (2000), J Mol Biol, 303, 531-543. Structures of l-fuculose-1-phosphate aldolase mutants outlining motions during catalysis. DOI:10.1006/jmbi.2000.4153. PMID:11054289.
- Dreyer MK et al. (1996), J Mol Biol, 259, 458-466. Catalytic Mechanism of the Metal-dependent Fuculose Aldolase fromEscherichia colias Derived from the Structure. DOI:10.1006/jmbi.1996.0332. PMID:8676381.
- Dreyer MK et al. (1996), Acta Crystallogr D Biol Crystallogr, 52, 1082-1091. Refined High-Resolution Structure of the Metal-Ion Dependent L-Fuculose-1-phosphate Aldolase (Class II) from Escherichia coli. DOI:10.1107/s0907444996009146. PMID:15299567.
- Dreyer MK et al. (1993), J Mol Biol, 231, 549-553. The Spatial Structure of the Class II l-Fuculose-1-phosphate Aldolase from Escherichia coli. DOI:10.1006/jmbi.1993.1307. PMID:8515438.
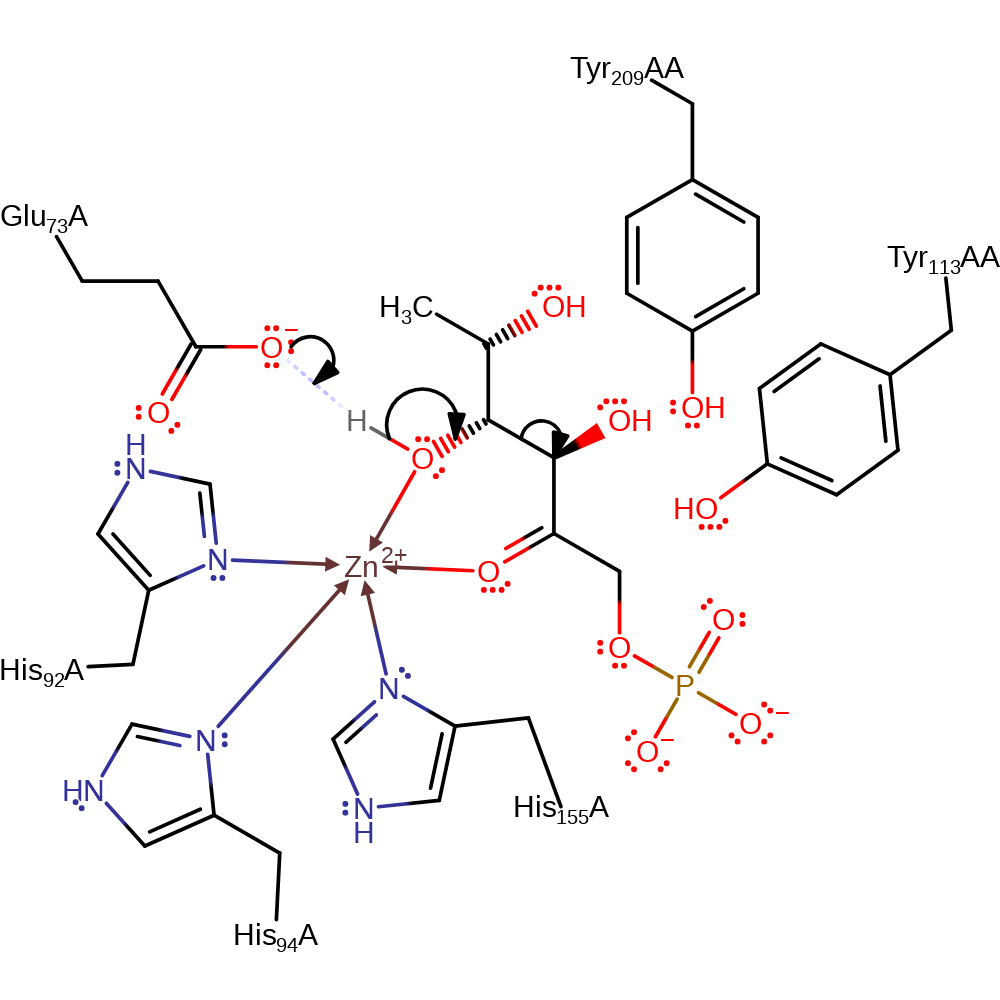
Step 1. It is assumed that the enzyme binds the open form of the sugar. Glu73 deprotonates the substrate alcohol at the C4 position, resulting in the elimination of the ene-diolate intermediate and the formation of the product lactaldehyde.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu73A | hydrogen bond acceptor |
| His92A | metal ligand |
| His155A | metal ligand |
| His94A | metal ligand |
| Tyr113A(AB) | transition state stabiliser |
| Tyr209A(AB) | transition state stabiliser |
| Glu73A | proton acceptor |
Chemical Components
ingold: bimolecular elimination, overall reactant used, overall product formed, intermediate formationCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu73A | hydrogen bond donor |
| Tyr113A(AB) | electrostatic stabiliser |
| His92A | metal ligand |
| His155A | metal ligand |
| His94A | metal ligand |
| Tyr209A(AB) | electrostatic stabiliser |
| Glu73A | proton donor |



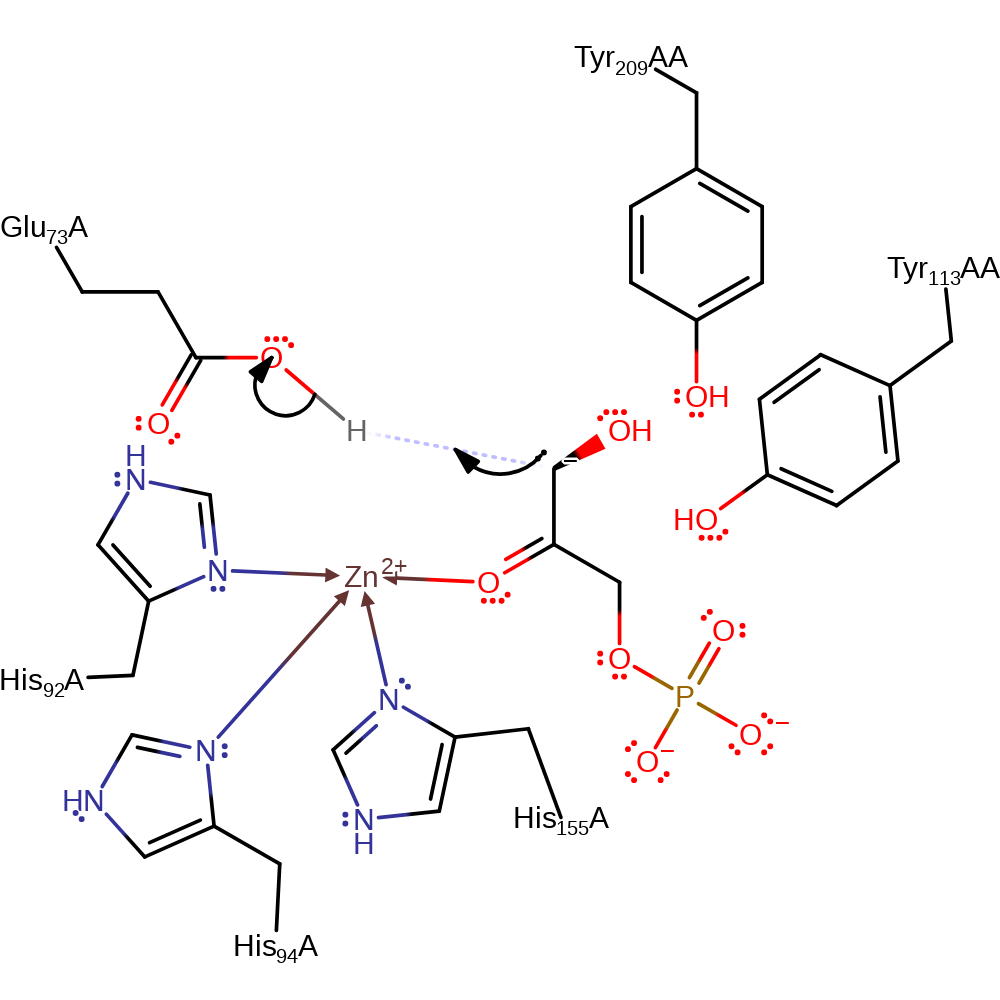
 Download:
Download: