Ornithine cyclodeaminase
Ornithine cyclodeaminase (OCD) from Pseudomonas putida belongs to the mu-crystallin protein family. It catalyses the conversion of L-ornithine to L-proline by an NAD+ dependent hydride transfer reaction. The precise function of this protein is still unknown, but it may have a regulatory function in the use of amino acids as neurotransmitters.
Reference Protein and Structure
- Sequence
-
Q88H32
 (4.3.1.12)
(4.3.1.12)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Pseudomonas putida KT2440 (Bacteria)

- PDB
-
1x7d
- Crystal Structure Analysis of Ornithine Cyclodeaminase Complexed with NAD and ornithine to 1.6 Angstroms
(1.6 Å)



- Catalytic CATH Domains
-
3.30.1780.10
 3.40.50.720
3.40.50.720  (see all for 1x7d)
(see all for 1x7d)
- Cofactors
- Nadph(4-) (1)
Enzyme Reaction (EC:4.3.1.12)
Enzyme Mechanism
Introduction
Asp 228 abstracts a proton from the alpha-amino nitrogen, which causes a hydride to be transferred from the alpha-carbon to the re face of NAD+. Glu 56 acts as a general base by deprotonating the nitrogen atom of the delta-amino group, activating it for nucleophilic attack on the alpha-carbon atom. This results in the double bond between the alpha-carbon and nitrogen breaking, and the alpha-nitrogen being protonated by Asp 228. Asp 228 then deprotonates the delta-nitrogen, causing it to nucleophilically attack the alpha-carbon again, forming a double bond and eliminating ammonia. This results in an electrophilic C2 centre which then accepts a hydride from NADH, breaking the C=N to form L-proline and reforming NAD+.
Catalytic Residues Roles
| UniProt | PDB* (1x7d) | ||
| Glu56 | Glu56A | Acts as a general base, deprotonating the nitrogen atom of the delta-amino group, activating it for nucleophilic attack on the alpha-carbon. | increase nucleophilicity, proton acceptor, proton donor |
| Asp228 | Asp228A | Acts as an acid/base to the substrate by deprotonating the alpha-amino nitrogen, activating it for nucleophilic attack. It then donates this proton back to the alpha-nitrogen once nucleophilic attack has taken place. Asp 228 then deprotonates the delta-nitrogen, activating it for nucleophilic attack. | proton acceptor, proton donor |
Chemical Components
proton transfer, hydride transfer, cofactor used, overall reactant used, intramolecular nucleophilic addition, cyclisation, bimolecular elimination, native state of cofactor regenerated, overall product formed, bimolecular nucleophilic additionReferences
- Goodman JL et al. (2004), Biochemistry, 43, 13883-13891. Ornithine Cyclodeaminase: Structure, Mechanism of Action, and Implications for the μ-Crystallin Family†,‡. DOI:10.1021/bi048207i. PMID:15518536.
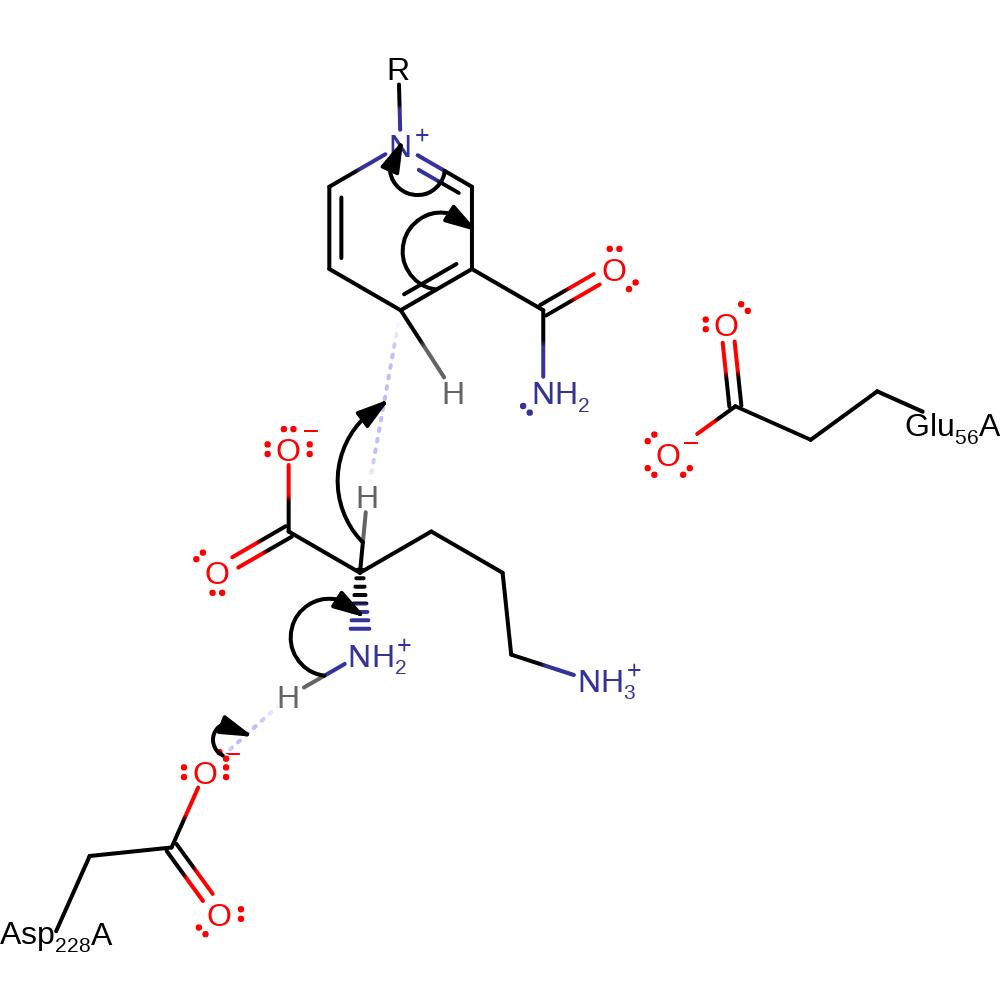
Step 1. Asp 228 abstracts a proton from the alpha-amino nitrogen, which causes a hydride to be transferred from the alpha-carbon to the re face of NAD+
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp228A | proton acceptor |
Chemical Components
proton transfer, hydride transfer, cofactor used, overall reactant used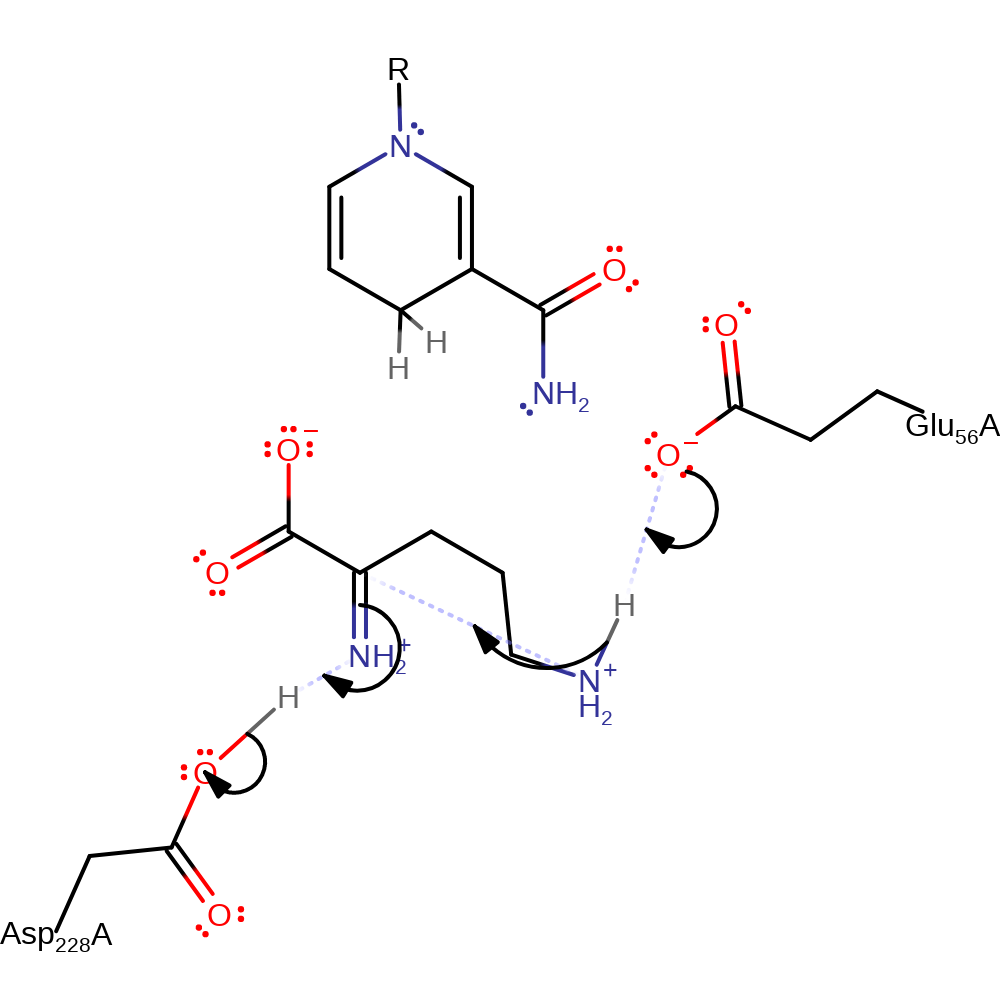
Step 2. Glu 56 acts as a general base by deprotonating the nitrogen atom of the delta-amino group, activating it for nucleophilic attack on the alpha-carbon atom. This results in the double bond between the alpha-carbon and nitrogen breaking, and the alpha-nitrogen being protonated by Asp 228.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu56A | increase nucleophilicity |
| Asp228A | proton donor |
| Glu56A | proton acceptor |
Chemical Components
proton transfer, ingold: intramolecular nucleophilic addition, cyclisation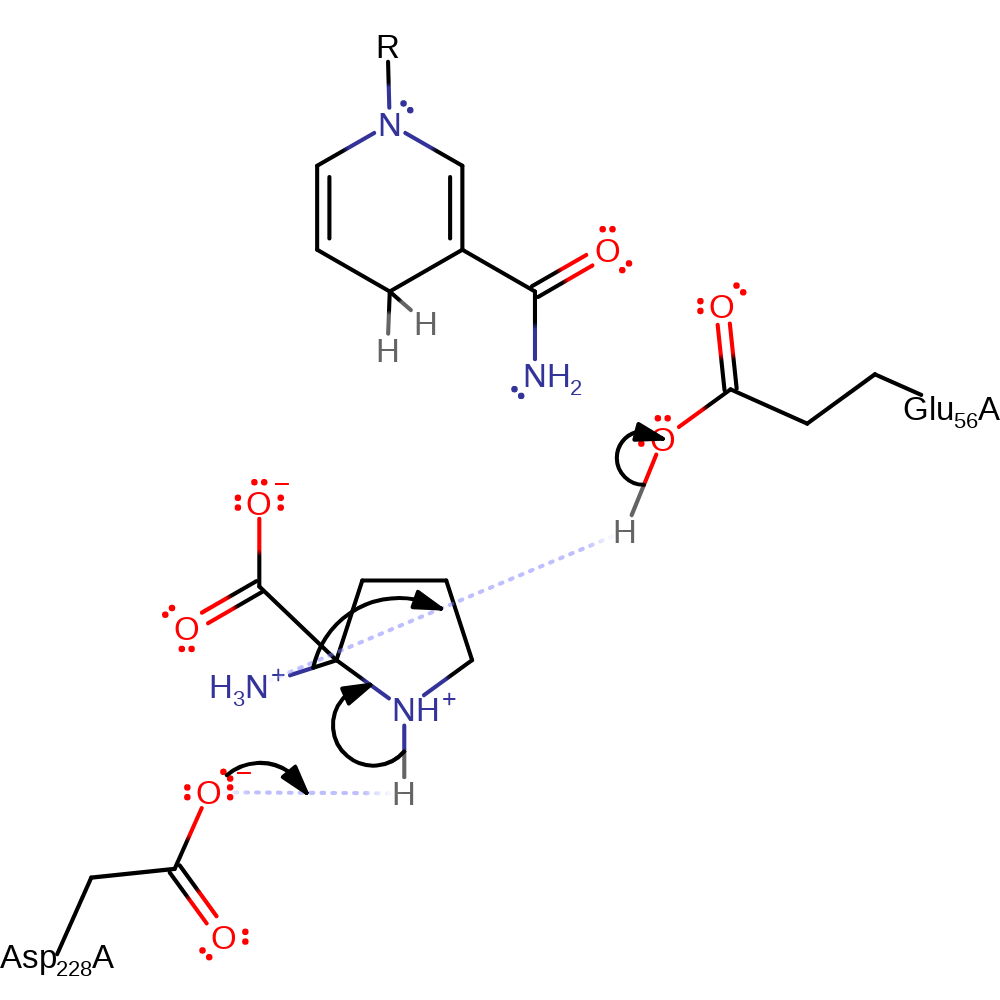
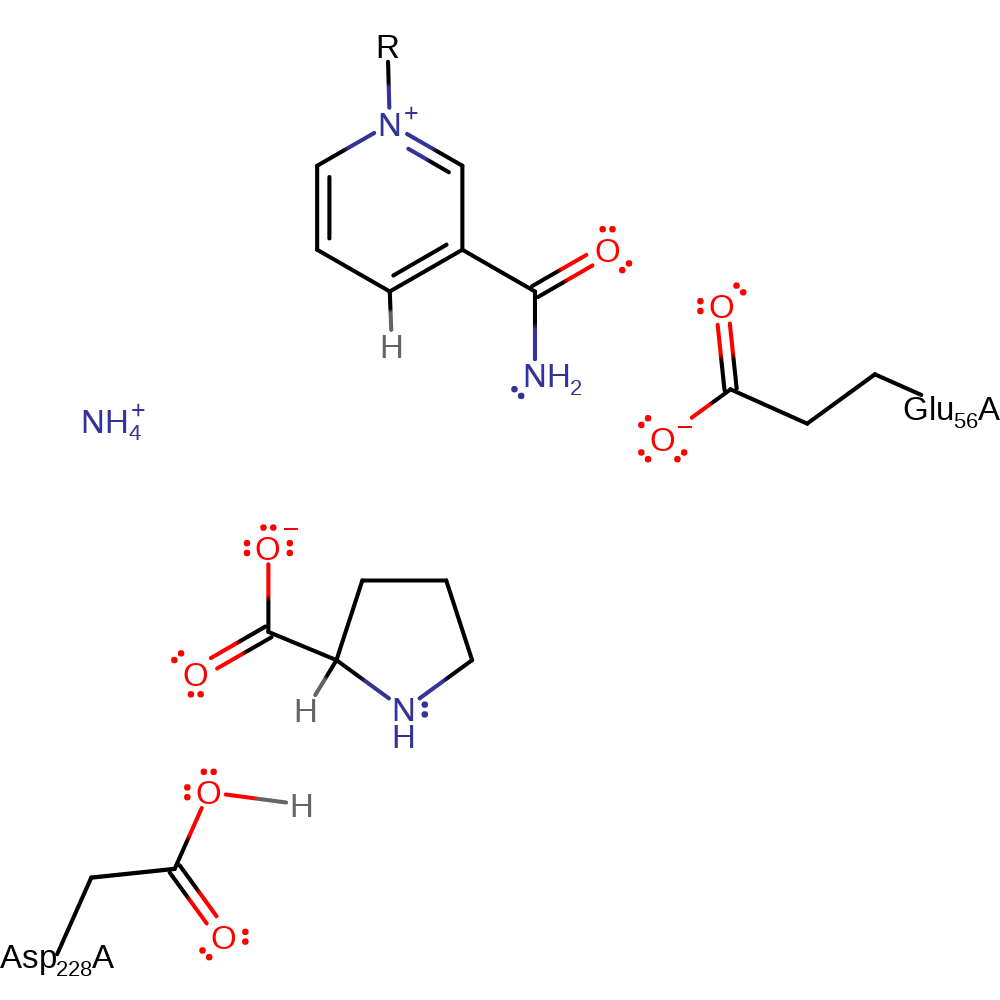
Step 3. Asp 228, which loses a proton to an unknown base between the previous step and this one then deprotonates the delta-nitrogen, causing it to nucleophilically attack the alpha-carbon again, forming a double bond and eliminating ammonia.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp228A | proton acceptor |
| Glu56A | proton donor |
Chemical Components
ingold: bimolecular elimination, proton transfer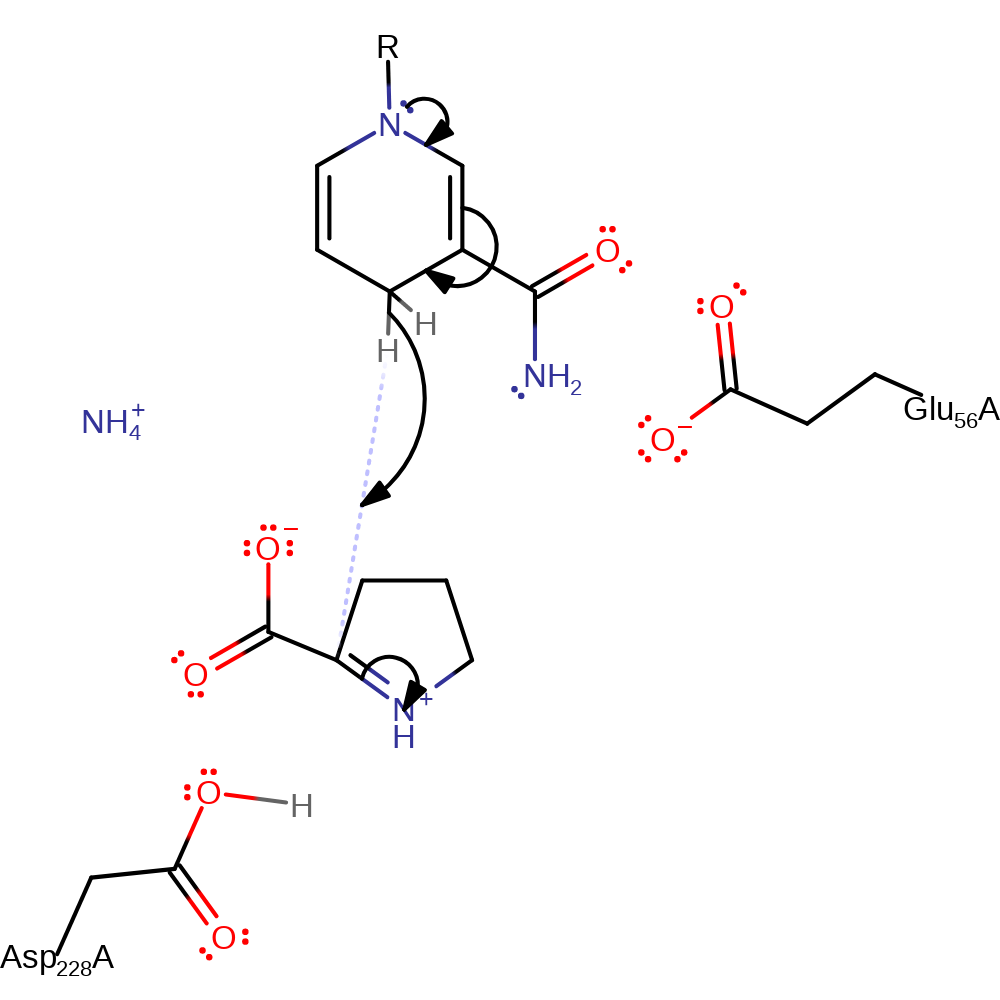
Step 4. The electrophilic C2 centre then accepts a hydride from NADH, breaking the C=N to form L-proline and reforming NAD+.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|



 Download:
Download: