Glyceraldehyde-3-phosphate dehydrogenase (NADP+)
NADP-dependent glyceraldehyde-3-phosphate dehydrogenase (GAPN) from Streptococcus mutans (GAPN) is a non-phosphorylating, CoA-independent member of the aldehyde dehydrogenase (ALDH) family. It catalyses the irreversible oxidation of glyceraldehyde-3-phosphate (G3P) into 3-phosphoglycerate (3-PGA) in the presence of NADP via a two step mechanism.
Reference Protein and Structure
- Sequence
-
Q59931
 (1.2.1.9)
(1.2.1.9)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Streptococcus mutans UA159 (Bacteria)

- PDB
-
2esd
- Crystal Structure of thioacylenzyme intermediate of an Nadp Dependent Aldehyde Dehydrogenase
(2.55 Å)



- Catalytic CATH Domains
-
3.40.309.10
 3.40.605.10
3.40.605.10  (see all for 2esd)
(see all for 2esd)
- Cofactors
- Nadp zwitterion (1)
Enzyme Reaction (EC:1.2.1.9)
Enzyme Mechanism
Introduction
GAPN uses an ordered sequential two-step mechanism for catalysis - acylation followed by deacylation. First, NADP+ binds to the enzyme, inducing a conformational change in the active site, resulting in the activation of a competent enzyme. The positive charge of the nicotinamide ring of NADP+, the amide peptide nitrogens of residues Cys284 and Thr285, and possibly the involvement of other structural elements stabilise the thiolate form of Cys284. G3P binding leads to formation of a thiohemiacetal intermediate via a nucleophilic attack by the Cys284 thiolate. This is followed by an oxidoreduction process in which a hydride is transferred from the intermediate to the nicotinamide ring of NADP+ forming a thioacylenzyme intermediate. This is able to take place without catalytic assistance due to the presence of an oxianion hole, formed by Asn154 and the amide nitrogen of Cys284, interacting with the C1 oxygen of the substrate, keeping the oxygen deprotonated. The oxyanion hole is also involved in the stabilisation of the tetrahedral thioacyl intermediate. Then deacylation occurs, where Glu250 orientates and activates water which nucleophilically attacks the C1 carbon of the thioacyl intermediate, leading to an acid product and, finally, the release of the reduced cofactor (NADPH). The isomerisation and movement of the nicotinamide ring of NADPH after the oxidoreduction stage is required to allow Glu250 to perform its role in deacylation.
Catalytic Residues Roles
| UniProt | PDB* (2esd) | ||
| Glu250 | Ala250A | Glu250 orientates and activates water which acts as a nucleophile and attacks the C1 carbon of the thioacylenzyme intermediate, leading to the formation an acid product of 3-PGA and the release of the reduced cofactor. At basic pH, deacylation is catalysed by the chemical activation of the water molecule attacking the thioacyl intermediate via the abstraction of a proton by Glu250 acting as a base catalyst. | proton acceptor, activator, electrostatic stabiliser, proton donor |
| Asn154 | Asn154A | Asn154-ND2 group and the amide nitrogen of Cys284 interact with the C1 oxygen of the substrate to form an oxianion hole which is required of hydride transfer and involved in the stabilisation of the intermediates. | electrostatic stabiliser |
| Cys284 | Cys284A | In thiolate form, Cys284 acts as a nucleophile and attacks C1 of G3P substrate to form a thiohemiacetal enzyme intermediate in the acylation process. The amide nitrogen of Cys284, in conjunction with Asn154, interacts with the C1 oxygen of the substrate to form an oxianion hole which is required for hydride transfer and involved in the stabilisation of the intermediates. | covalently attached, nucleofuge, nucleophile, proton acceptor, electrostatic stabiliser |
Chemical Components
bimolecular nucleophilic addition, intermediate formation, overall reactant used, enzyme-substrate complex formation, aromatic bimolecular nucleophilic addition, hydride transfer, intermediate collapse, cofactor used, proton transfer, overall product formed, hydrolysis, unimolecular elimination by the conjugate base, native state of enzyme regenerated, inferred reaction stepReferences
- D'Ambrosio K et al. (2006), Biochemistry, 45, 2978-2986. The First Crystal Structure of a Thioacylenzyme Intermediate in the ALDH Family: New Coenzyme Conformation and Relevance to Catalysis†. DOI:10.1021/bi0515117. PMID:16503652.
- Marchal S et al. (2001), Chem Biol Interact, 130-132, 15-28. Chemical mechanism and substrate binding sites of NADP-dependent aldehyde dehydrogenase from Streptococcus mutans. DOI:10.1016/s0009-2797(00)00218-0. PMID:11306027.
- Marchal S et al. (2000), Biochemistry, 39, 3327-3335. Role of Glutamate-268 in the Catalytic Mechanism of Nonphosphorylating Glyceraldehyde-3-phosphate Dehydrogenase fromStreptococcus mutans†. DOI:10.1021/bi9914208. PMID:10727225.
- Cobessi D et al. (2000), J Mol Biol, 300, 141-152. Structural and biochemical investigations of the catalytic mechanism of an NADP-dependent aldehyde dehydrogenase from Streptococcus mutans. DOI:10.1006/jmbi.2000.3824. PMID:10864505.
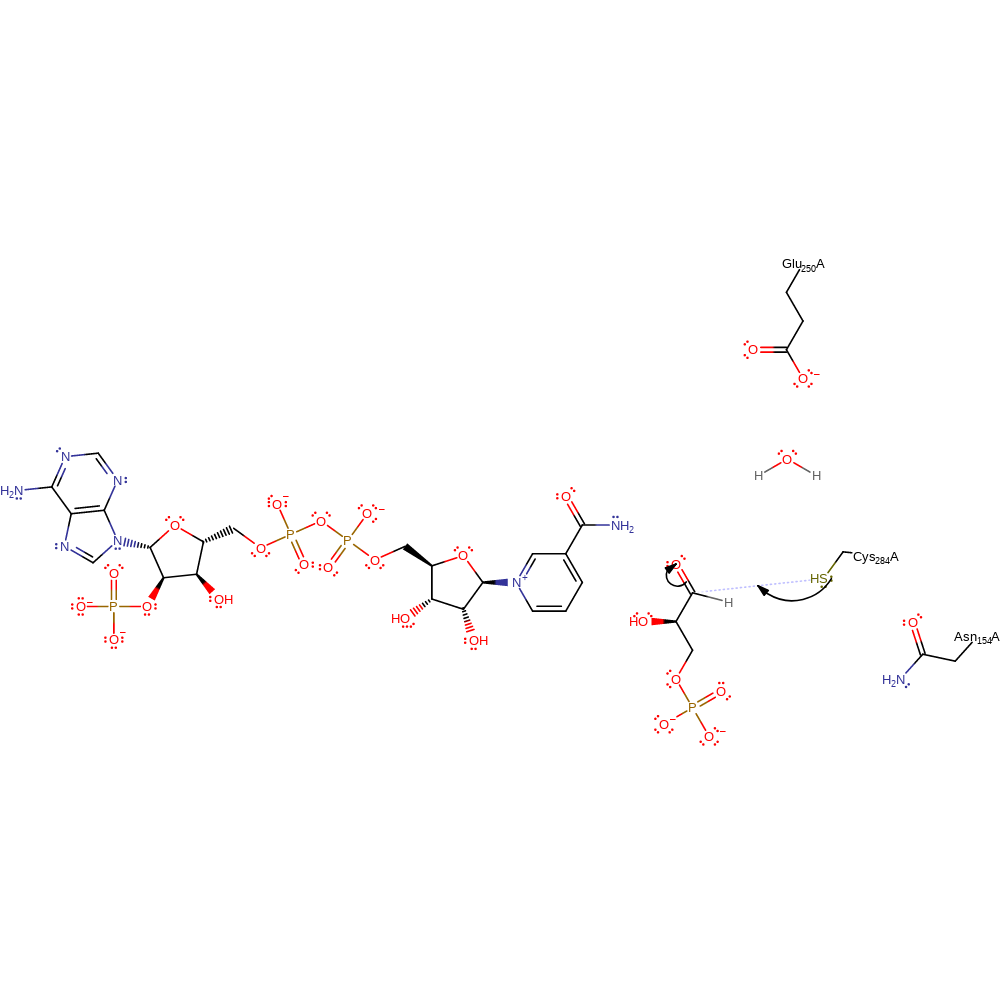
Step 1. The thiolate of Cys284 attacks G3P in a nucleophilic addition reaction.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn154A | electrostatic stabiliser |
| Cys284A | nucleophile |
Chemical Components
ingold: bimolecular nucleophilic addition, intermediate formation, overall reactant used, enzyme-substrate complex formation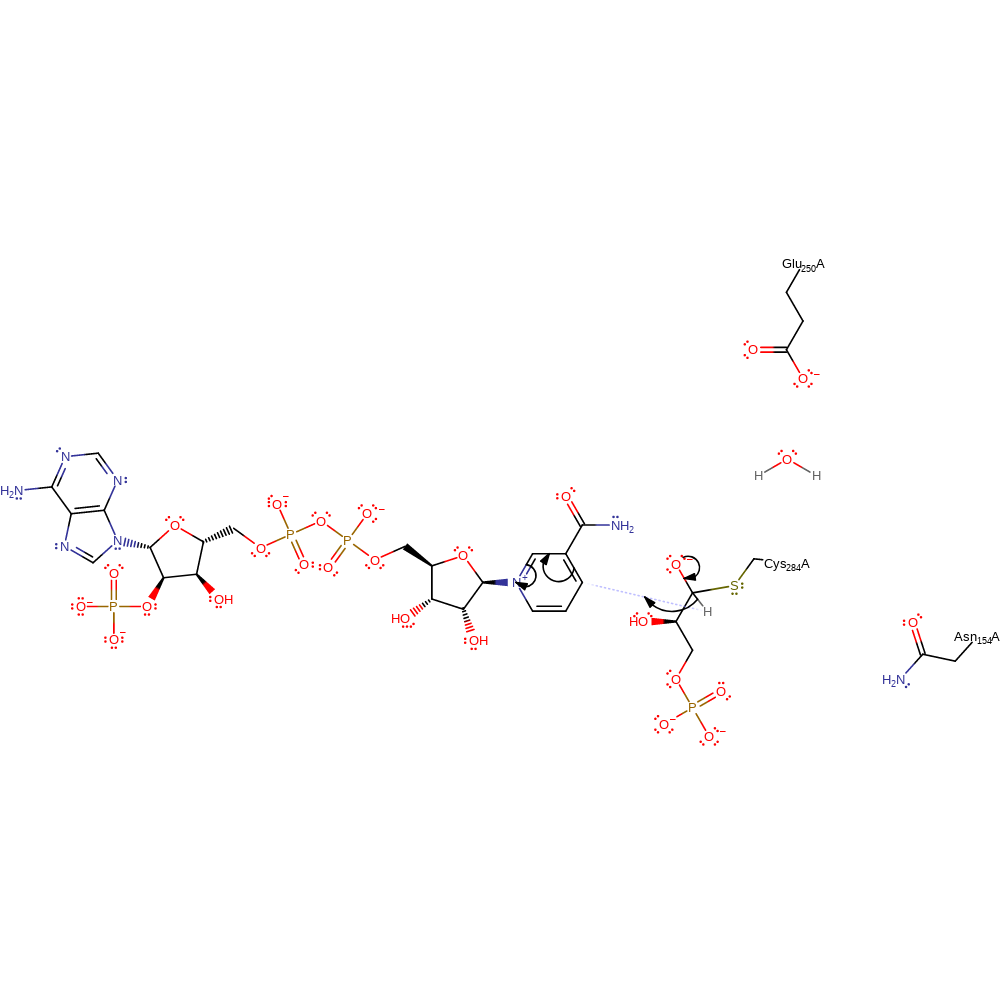
Step 2. The tetrahedral intermediate collapses and a hydride is transferred to NADP.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Cys284A | covalently attached |
| Asn154A | electrostatic stabiliser |
| Cys284A | electrostatic stabiliser |
Chemical Components
ingold: aromatic bimolecular nucleophilic addition, hydride transfer, intermediate collapse, cofactor used
Step 3. Glu250 activates a water molecule for nucleophilic attack of the intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Ala250A | activator |
| Asn154A | electrostatic stabiliser |
| Ala250A | electrostatic stabiliser |
| Cys284A | covalently attached |
| Ala250A | proton acceptor |
Chemical Components
ingold: bimolecular nucleophilic addition, proton transfer, intermediate formation
Step 4. The tetrahedral intermediate collapses, eliminating Cys and forming the product.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn154A | electrostatic stabiliser |
| Ala250A | electrostatic stabiliser |
| Cys284A | nucleofuge |
Chemical Components
overall product formed, hydrolysis, ingold: unimolecular elimination by the conjugate base
Step 5. In an inferred step the native state of the enzyme is restored when Cys284 is protonated and Glu250 is deprotonated.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Ala250A | proton donor |
| Cys284A | proton acceptor |






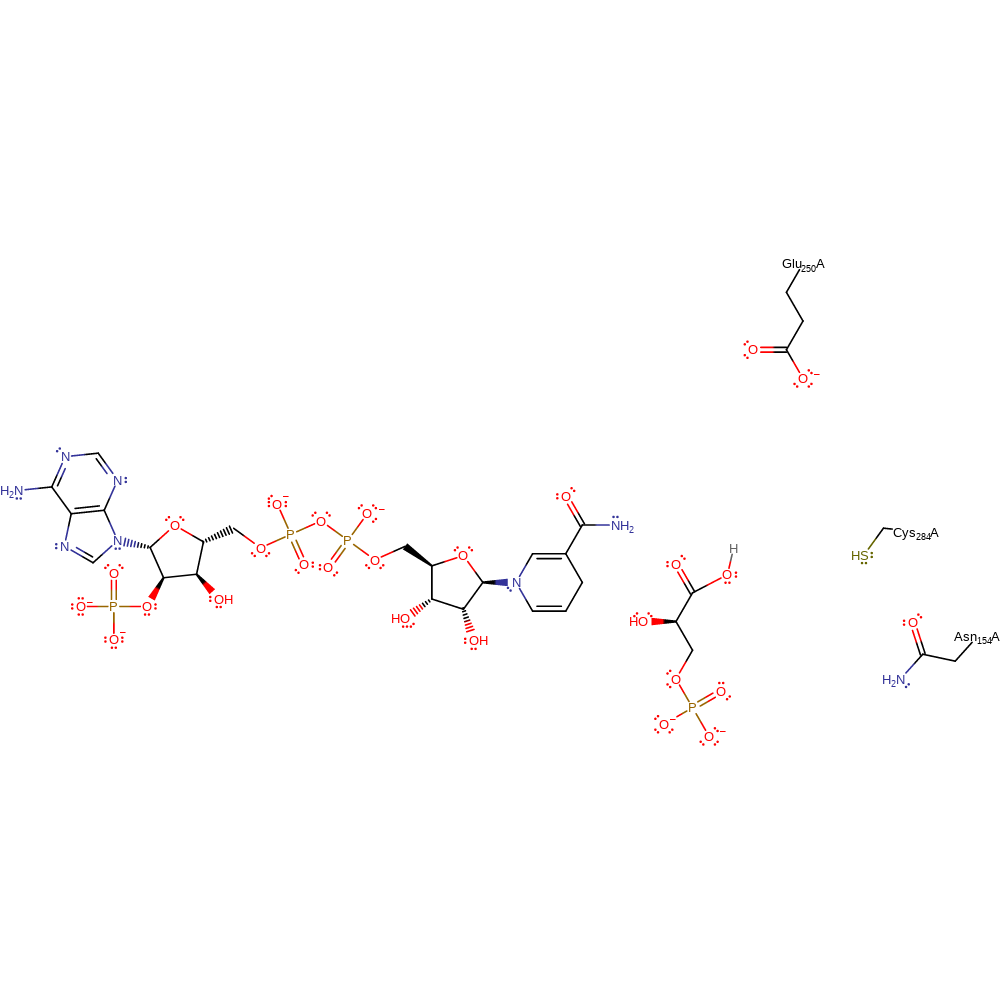 Download:
Download: