Cytosine deaminase (bacterial)
Cytosine deaminase (CD) catalyses the deamination of cytosine to uracil and ammonia. It is an enzyme produced by bacteria and yeast, but not mammalian cells, and is a potential gene therapy agent. CD deaminates 5-fluorocytosine to potent antimetabolite 5-fluorouracil, which can block DNA replication in cells. Bacterial CD (bCD) is capable of deaminating a wide range of cytosine derivatives with varying efficiency. It is dependent of Fe2+ for maximal catalytic activity.
Reference Protein and Structure
- Sequence
-
P25524
 (3.5.4.-, 3.5.4.1)
(3.5.4.-, 3.5.4.1)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Escherichia coli K-12 (Bacteria)

- PDB
-
1ra0
- Bacterial cytosine deaminase D314G mutant bound to 5-fluoro-4-(S)-hydroxy-3,4-dihydropyrimidine.
(1.12 Å)



- Catalytic CATH Domains
-
3.20.20.140
 (see all for 1ra0)
(see all for 1ra0)
- Cofactors
- Zinc(2+) (1)
Enzyme Reaction (EC:3.5.4.1)
Enzyme Mechanism
Introduction
Glu217 initially deprotonates the Zn bound and then after some structural movements transfers the proton to the N3 nitrogen. The hydroxyl can then attack the C4 carbon and then the amino group accepts a proton from the hydroxyl which initiates the collapse of the tetrahedral intermediate which releases the products: ammonia and uracil.
Catalytic Residues Roles
| UniProt | PDB* (1ra0) | ||
| Asp314, His62, His64, His215 | Asp313(317)A, His61(65)A, His63(67)A, His214(218)A | Bind the metal ion. | metal ligand |
| Glu218 | Glu217(221)A | Glu217 acts as a catalytic acid/base residue. It donates a proton to the N3 nitrogen, increasing the susceptibility of the C4 carbon to nucleophilic attack by weakening the N3 to C4 double bond character. | proton acceptor, proton donor |
| Gln157 | Gln156(160)A | Gln156 forms hydrogen bonds with the N1 nitrogen and O2 oxygen of the pyrimidine ring, helping to orientate the substrate, and polarise the aromatic structure of the pyrimidine ring, also making the C4 carbon more reactive towards an addition by the activated water molecule. | electrostatic stabiliser |
Chemical Components
proton transfer, intermediate formation, overall reactant used, bimolecular nucleophilic addition, cofactor used, unimolecular elimination by the conjugate base, decoordination from a metal ion, heterolysis, intermediate collapse, native state of enzyme regenerated, overall product formedReferences
- Ireton GC et al. (2002), J Mol Biol, 315, 687-697. The structure of Escherichia coli cytosine deaminase. DOI:10.1006/jmbi.2001.5277. PMID:11812140.
- Manta B et al. (2014), J Phys Chem B, 118, 5644-5652. Reaction mechanism of zinc-dependent cytosine deaminase from Escherichia coli: a quantum-chemical study. DOI:10.1021/jp501228s. PMID:24833316.
- Mahan SD et al. (2004), Protein Eng Des Sel, 17, 625-633. Random mutagenesis and selection of Escherichia coli cytosine deaminase for cancer gene therapy. DOI:10.1093/protein/gzh074. PMID:15381761.
- Porter DJ et al. (1993), J Biol Chem, 268, 24005-24011. Cytosine deaminase. The roles of divalent metal ions in catalysis. PMID:8226944.
Catalytic Residues Roles
| Residue | Roles |
|---|---|
| Gln156(160)A | electrostatic stabiliser |
| His61(65)A | metal ligand |
| His63(67)A | metal ligand |
| His214(218)A | metal ligand |
| Asp313(317)A | metal ligand |
| Glu217(221)A | proton acceptor |
Chemical Components
proton transfer, intermediate formation, overall reactant used
Step 2. Glu217 has to break its hydrogen bond to the nucleophile, rotate toward the substrate, and create a new hydrogen bond to the N3 nitrogen where it transfers a proton to the N3 nitrogen.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gln156(160)A | electrostatic stabiliser |
| His61(65)A | metal ligand |
| His63(67)A | metal ligand |
| His214(218)A | metal ligand |
| Asp313(317)A | metal ligand |
| Glu217(221)A | proton donor |
Chemical Components
proton transfer, intermediate formation
Step 3. The Zinc bound hydroxyl nucleophilically attacks the C4 carbon which is more electrophilic due to the protonation at N3.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gln156(160)A | electrostatic stabiliser |
| His61(65)A | metal ligand |
| His63(67)A | metal ligand |
| His214(218)A | metal ligand |
| Asp313(317)A | metal ligand |
Chemical Components
ingold: bimolecular nucleophilic addition, cofactor used, intermediate formation
Step 4. The amino group bound to C4 accepts a proton from the hydroxyl group.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gln156(160)A | electrostatic stabiliser |
| His61(65)A | metal ligand |
| His63(67)A | metal ligand |
| His214(218)A | metal ligand |
| Asp313(317)A | metal ligand |
Chemical Components
proton transfer, intermediate formation
Step 5. The protonation of the amino group results in the initiation of an elimination from the oxyanion which leads to the release of ammonia and uracil.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gln156(160)A | electrostatic stabiliser |
| His61(65)A | metal ligand |
| His63(67)A | metal ligand |
| His214(218)A | metal ligand |
| Asp313(317)A | metal ligand |


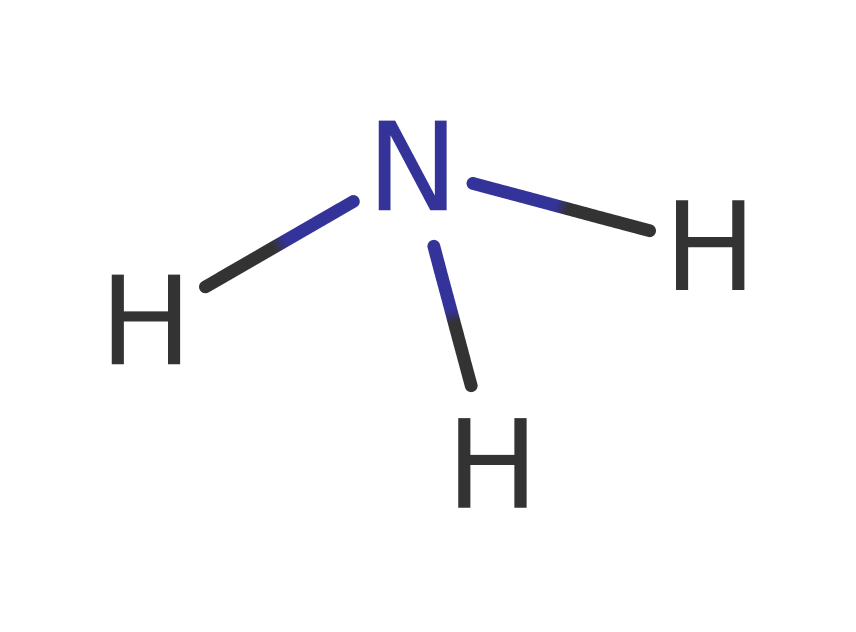
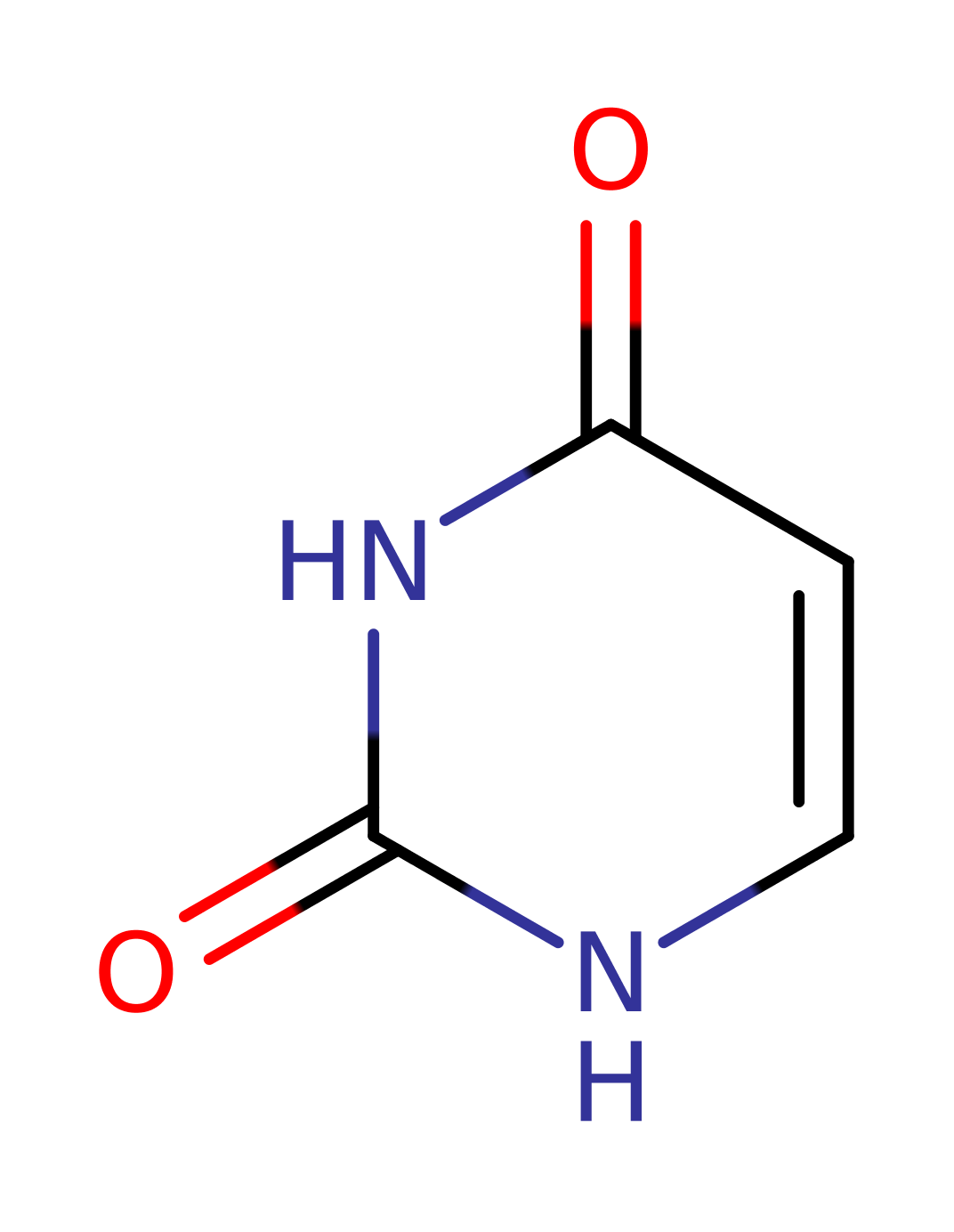

 Download:
Download: