Protein-L-isoaspartate(D-aspartate) O-methyltransferase
Formation of isoaspartyl residues is one of several processes that damage proteins as they age. Protein L-isoaspartate (D-aspartate) O-methyltransferase (PIMT) is a conserved and nearly ubiquitous enzyme that catalyzes the repair of proteins damaged by isoaspartyl formation.
Reference Protein and Structure
- Sequence
-
Q56308
 (2.1.1.77)
(2.1.1.77)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Thermotoga maritima MSB8 (Bacteria)

- PDB
-
1dl5
- PROTEIN-L-ISOASPARTATE O-METHYLTRANSFERASE
(1.8 Å)



- Catalytic CATH Domains
-
3.40.50.150
 (see all for 1dl5)
(see all for 1dl5)
Enzyme Reaction (EC:2.1.1.77)
Enzyme Mechanism
Introduction
PIMT catalyses the transfer of a methyl group from S-adenosyl-L-methionine (SAM) to the alpha-carboxylate side chain of the isoaspartyl residue. Through this transfer, the isoaspartyl group becomes activated for conversion back to the succinimide via a nonenzymatic attack of the nitrogen lone pair of the beta-peptide group on the alpha-carboxylate group. Once back in the succinimide form, hydrolysis can again occur at either carbonyl carbon, converting the protein to the aspartate form or regenerating the isoaspartate. This process cannot restore the amino group to the side chain of an original asparagine, but the pathway can restore the correct configuration to the protein backbone.
Catalytic Residues Roles
| UniProt | PDB* (1dl5) | ||
| Ser59 | Ser59A | Hydrogen bonds to the alpha-carboxylate group promoting the in line attack of O1 on SAM. | electrostatic stabiliser |
Chemical Components
bimolecular nucleophilic substitution, overall product formed, overall reactant used, cofactor usedReferences
- Skinner MM et al. (2000), Structure, 8, 1189-1201. Crystal structure of protein isoaspartyl methyltransferase: a catalyst for protein repair. PMID:11080641.
- Bennett EJ et al. (2003), Biochemistry, 42, 12844-12853. Catalytic implications from the Drosophila protein L-isoaspartyl methyltransferase structure and site-directed mutagenesis. DOI:10.1021/bi034891. PMID:14596598.
- Griffith SC et al. (2001), J Mol Biol, 313, 1103-1116. Crystal structure of a protein repair methyltransferase from Pyrococcus furiosus with its l-isoaspartyl peptide substrate. DOI:10.1006/jmbi.2001.5095. PMID:11700066.
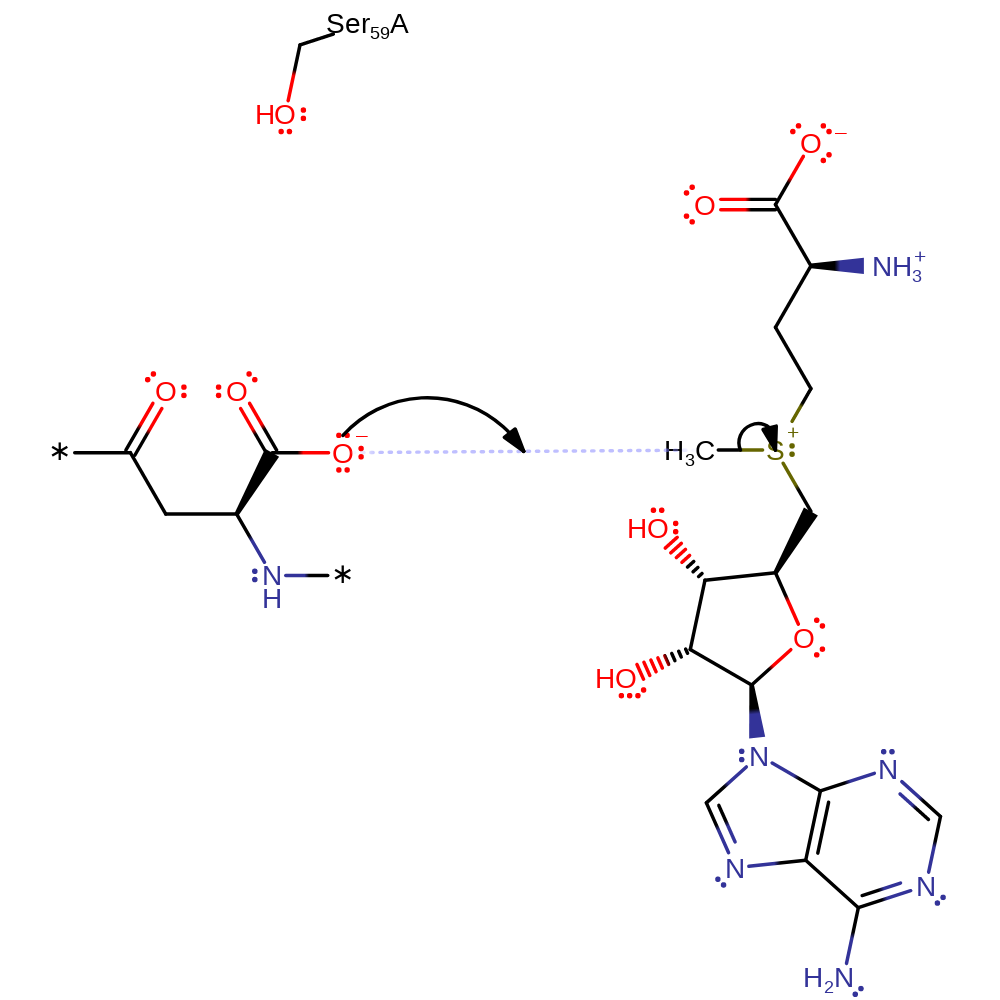
Step 1. In an SN2 reaction the carboxylate oxygen of the isoaspartyl group attacks the methyl of SAM. This forms the product and SAH.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Ser59A | electrostatic stabiliser |




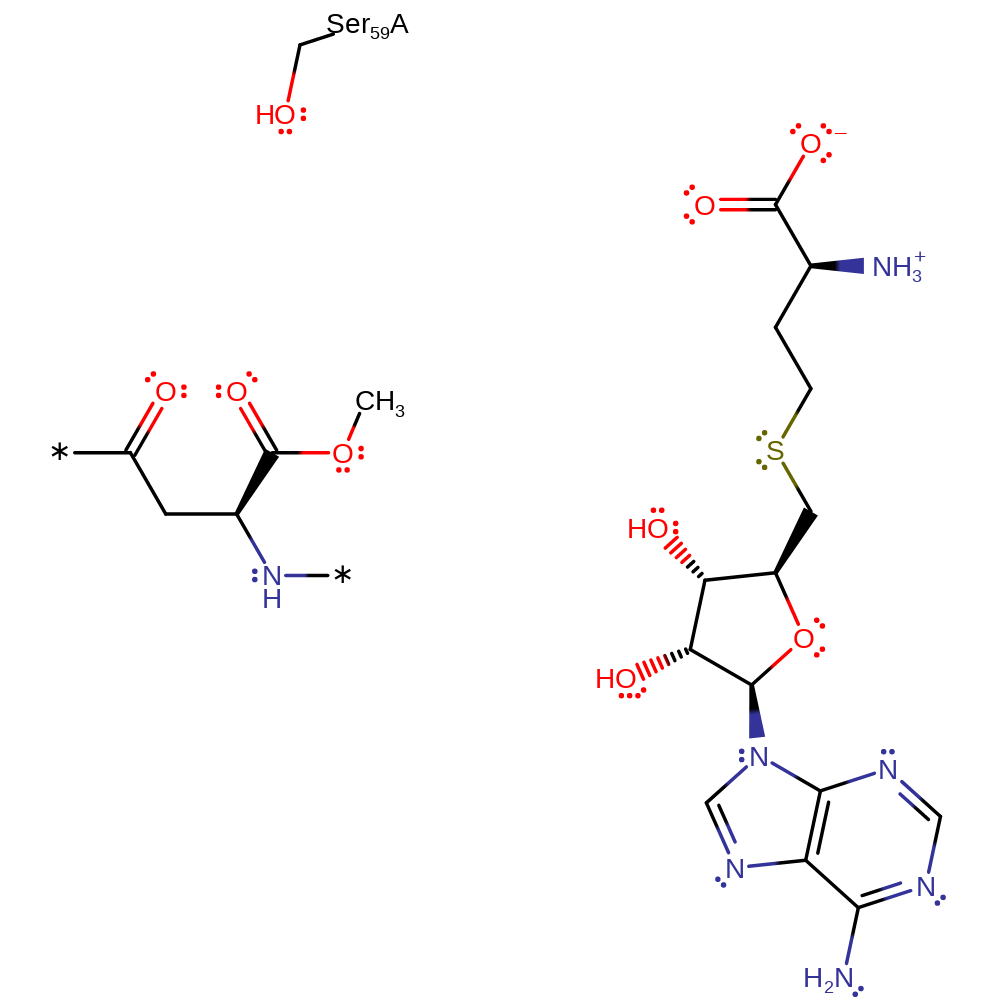 Download:
Download: