D-stereospecific aminopeptidase (peptidase S58 family)
The L-aminopeptidase D-Ala-esterase/amidase from Ochrobactrum anthropi (DmpA) is an aminopeptidase which catalyses the release of N-terminal residues from peptides substrates. It also shows D-amidasic and D-esterasic activities on D-alanine derivatives. DmpA and other β-aminopeptidases are able to to cleave synthetic β-peptides, which consist of backbone-elongated β-amino acid residues that are not processed by common proteolytic enzymes. β-peptides are considered promising building blocks for the design of novel peptidomimetics,small protein-like chains to mimic peptides which are useful for drug design, thus these enzymes that cleave and release them are useful for production of peptidominetics.
Reference Protein and Structure
- Sequence
-
Q59632
 (3.4.11.19)
(3.4.11.19)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Ochrobactrum anthropi (Bacteria)

- PDB
-
1b65
- Structure of l-aminopeptidase d-ala-esterase/amidase from ochrobactrum anthropi, a prototype for the serine aminopeptidases, reveals a new variant among the ntn hydrolase fold
(1.82 Å)



- Catalytic CATH Domains
-
3.60.70.12
 (see all for 1b65)
(see all for 1b65)
Enzyme Mechanism
Introduction
Based on the homologies of the catalytic centres between DmpA and Ntn hydrolases, the catalytic mechanism of DmpA was proposed to follow that of Ntn hydrolases. Ser250 alpha-amino group acts as a base to deprotonate its own hydroxyl group, which nucleophilically attacks the carbonyl group of the substrate. This results in the formation of a covalent enzyme-substrate transition state, stabilised by the oxyanion hole formed by the mainchain nitrogen atom of Tyr146 and the side chain of Asn218. Alpha-amino group of Ser250 then protonates the leaving group and deprotonates a water molecule to allow it to restore the enzyme by a nucleophilic attack to the acylenzyme. Ser288 hydrogen bonds with the alpha-amino group of Ser250, altering its pKa to enhance its acid/base property. Backbone amide group of Gly289 hydrogen bonds with the hydroxyl group of Ser250 to increase its nucleophilicity.
Catalytic Residues Roles
| UniProt | PDB* (1b65) | ||
| Gly289 (main-N) | Gly289A (main-N) | Its backbone amide group hydrogen bonds with the hydroxyl group of Ser 250 to increase its nucleophilicity. | electrostatic stabiliser |
| Ser250 | Ser250A | Its alpha-amino group acts as a base to deprotonate its own hydroxyl group, which nucleophilically attacks the carbonyl group of the substrate to form a acylenzyme intermediate. Its alpha-amino group also protonates the leaving group and deprotonate a water molecule to restore the enzyme from the intermediate. | nucleofuge, nucleophile, proton acceptor, proton donor |
| Ser288 | Ser288A | It hydrogen bonds with the alpha-amino group of Ser 250, altering its pKa to enhance its acid/base property. | electrostatic stabiliser |
| Tyr146 (main-N) | Tyr146A (main-N) | Its backbone amide forms an oxyanion hole to stabilise the negatively charged transition state. | electrostatic stabiliser |
| Asn218 | Asn218A | It forms an oxyanion hole to stabilise the negatively charged transition state. | electrostatic stabiliser |
Chemical Components
proton transfer, bimolecular nucleophilic addition, intermediate formation, overall reactant used, unimolecular elimination by the conjugate base, intermediate collapse, overall product formed, native state of enzyme regeneratedReferences
- Bompard-Gilles C et al. (2000), Structure, 8, 153-162. A new variant of the Ntn hydrolase fold revealed by the crystal structure of l-aminopeptidase d-Ala-esterase/amidase from Ochrobactrum anthropi. DOI:10.1016/s0969-2126(00)00091-5. PMID:10673442.
- Merz T et al. (2012), Structure, 20, 1850-1860. Autoproteolytic and catalytic mechanisms for the β-aminopeptidase BapA--a member of the Ntn hydrolase family. DOI:10.1016/j.str.2012.07.017. PMID:22980995.
- Geueke B et al. (2006), FEBS J, 273, 5261-5272. Bacterial beta-peptidyl aminopeptidases with unique substrate specificities for beta-oligopeptides and mixed beta,alpha-oligopeptides. DOI:10.1111/j.1742-4658.2006.05519.x. PMID:17064315.

Step 1. The amine group of Ser250 deprotonates its own hydroxyl group activating it so it can attack the carbon of the carbonyl in a nucleophilic addition to produce an oxyanion intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn218A | electrostatic stabiliser |
| Gly289A (main-N) | electrostatic stabiliser |
| Tyr146A (main-N) | electrostatic stabiliser |
| Ser288A | electrostatic stabiliser |
| Ser250A | proton donor, nucleophile |
| Ser250A (main-N) | proton acceptor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, intermediate formation, overall reactant used
Step 2. The oxyanion initiates an elimination which results in the cleavage of the peptide bond. The N-terminal product accepts a proton from Ser250.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr146A (main-N) | electrostatic stabiliser |
| Asn218A | electrostatic stabiliser |
| Ser288A | electrostatic stabiliser |
| Gly289A (main-N) | electrostatic stabiliser |
| Ser250A (main-N) | proton donor |
Chemical Components
ingold: unimolecular elimination by the conjugate base, proton transfer, intermediate collapse, overall product formed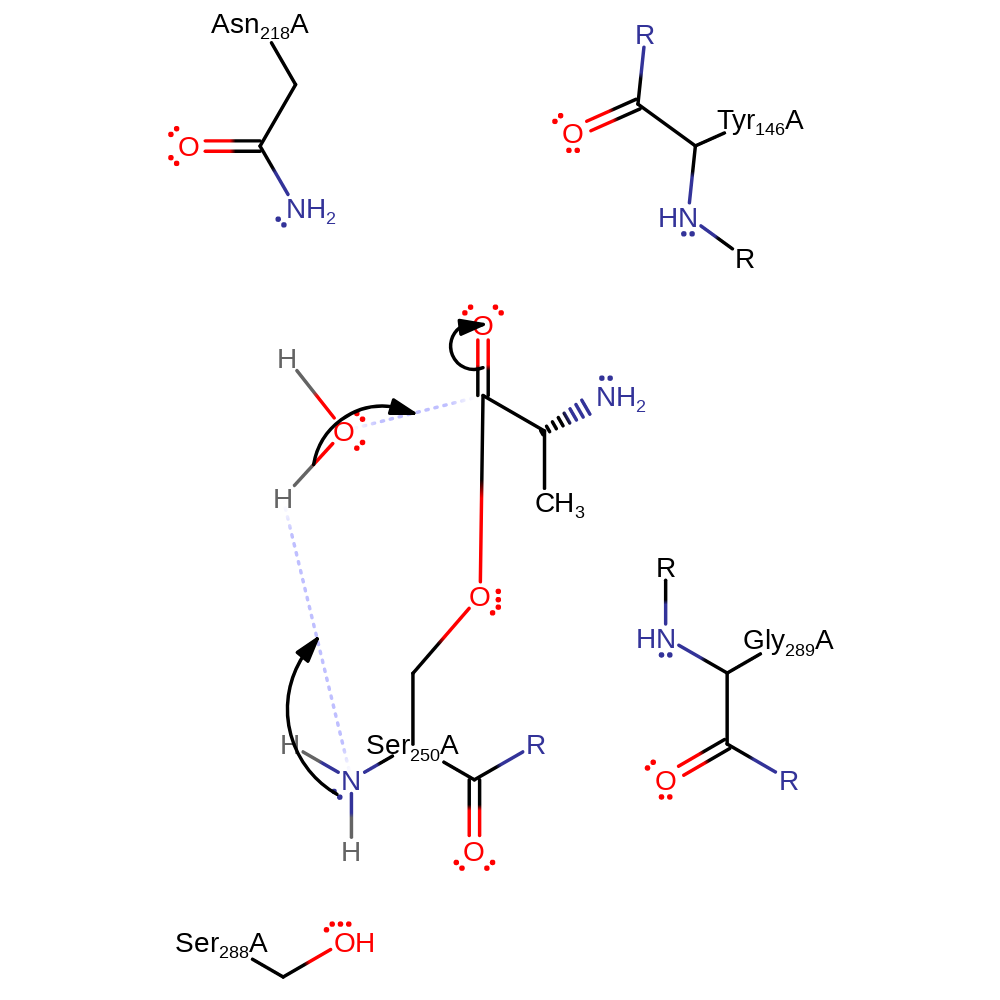
Step 3. The amine group of Ser250 abstracts a proton from a water which activates it so it can attack the carbon of the ester bond in a nucleophilic addition to produce another oxyanion intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr146A (main-N) | electrostatic stabiliser |
| Asn218A | electrostatic stabiliser |
| Ser288A | electrostatic stabiliser |
| Gly289A (main-N) | electrostatic stabiliser |
| Ser250A (main-N) | proton acceptor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, intermediate formation, overall reactant used
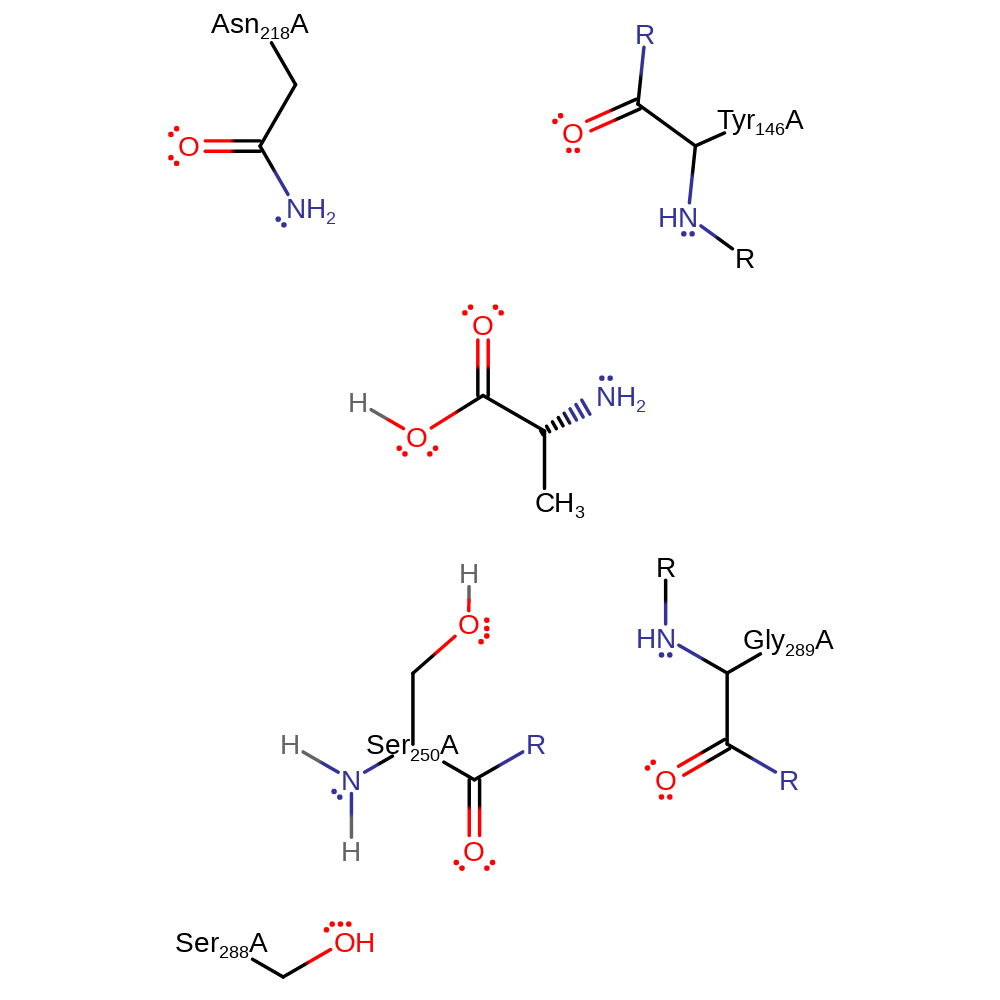
Step 4. The oxyanion initiates another elimination which results in the cleavage of the ester bond. The released Ser250 can now accepts a proton from its own amine group which returns the enzyme to its native state.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr146A (main-N) | electrostatic stabiliser |
| Asn218A | electrostatic stabiliser |
| Ser288A | electrostatic stabiliser |
| Gly289A (main-N) | electrostatic stabiliser |
| Ser250A | proton acceptor |
| Ser250A (main-N) | proton donor |
| Ser250A | nucleofuge |



 Download:
Download: