Cellobiose dehydrogenase (acceptor)
Fungal cellobiose dehydrogenase (CDH) participates in the rotting of wood by catalysing a key step in the breakdown of cellulose (and lignin), namely the oxidation of cellobiose (a reaction product in cellulose hydrolysis) to cellobiono-1,5-lactone. This is used to provide a carbon source for the micro-organism. CDH is the only known extracellular flavocytochrome, a small and heterogeneous group of redox-active proteins that carry both haem and flavin prosthetic groups. Applications of CDH include degradation of various environmental pollutants.
It shows sequence and structural homology with glucose and cholesterol oxidase, two other known enzymes catalysing similar processes, so the mechanism is likely to be the same between the three. FAD is used as a cofactor for the reaction, being reduced by the substrate and subsequently reoxidised by a cytochromal haem group contained in the non-active site containing domain (PDB:1d7b)
Reference Protein and Structure
- Sequence
-
Q01738
 (1.1.99.18)
(1.1.99.18)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Phanerodontia chrysosporium

- PDB
-
1kdg
- Crystal structure of the flavin domain of cellobiose dehydrogenase
(1.5 Å)



- Catalytic CATH Domains
-
3.50.50.60
 3.30.410.10
3.30.410.10  (see all for 1kdg)
(see all for 1kdg)
- Cofactors
- 6-hydroxy-fad (1)
Enzyme Reaction (EC:1.1.99.18)
Enzyme Mechanism
Introduction
The generally agreed on mechanism for this reaction is that of the hydride transfer: His 689 acts as a general base by deprotonating the C1 hydroxyl group of the cellobiose substrate, causing hydride transfer from C1 of the substrate to FAD. Asn 732, by hydrogen bonding to O1 of the substrate, facilitates proton transfer to His 689. Tyr 609 hydrogen bonds to a water molecule, increasing the water's affinity for protons, and causing it to deprotonate His 689. This now leaves a reduced FAD (FADH2), the second proton most likely coming from a water molecule. The mechanism for the oxidation of FADH2 is speculated to be as follows: An internal electron transfer takes place, passing one electron from the flavin centre to the haem, reducing the haem group, and forming a flavin semiquinone. The reduced haem is oxidised by either Fe3+ (to form Fe2+) or superoxide O2- radical (to form hydrogen peroxide.) The semiquinone reacts with O2 to form the O2- radical, and returns to its initial oxidised state. Ferrous iron can then be oxidised by hydrogen peroxide to re-form Fe3+, and also OH- and an OH radical.
Catalytic Residues Roles
| UniProt | PDB* (1kdg) | ||
| His707 | His689(480)A | Acts as the general base to accept a proton from the anomeric carbon's OH group allowing the formation of the carbonyl. | proton acceptor, proton donor |
| Tyr627 | Tyr609(400)A | Hydrogen bonds to a water molecule, causing that water molecule to deprotonate the protonated His 689 residue. | activator, hydrogen bond donor |
| Asn750 | Asn732(523)A | Forms a hydrogen bond to the oxygen of the anomeric carbon's OH group thus assisting in the deprotonation at that site by His689. | enhance reactivity, electrostatic stabiliser |
Chemical Components
proton transfer, hydride transfer, overall product formed, overall reactant used, bimolecular nucleophilic addition, native state of enzyme regeneratedReferences
- Hallberg BM et al. (2003), J Biol Chem, 278, 7160-7166. Mechanism of the Reductive Half-reaction in Cellobiose Dehydrogenase. DOI:10.1074/jbc.m210961200. PMID:12493734.
- Hallberg BM et al. (2002), J Mol Biol, 315, 421-434. Crystal structure of the flavoprotein domain of the extracellular flavocytochrome cellobiose dehydrogenase. DOI:10.1006/jmbi.2001.5246. PMID:11786022.

Step 1. His689 acts as a general base and deprotonates the 1-hydroxyl group of the reactant. Simultaneously, a hydride is transferred from C1 to a nitrogen atom of the 6-hydroxy FAD in a nucleophilic addition reaction, forming the product and FADH-. Asn732 promotes deprotonation of the hydroxyl group by forming a hydrogen bond to it.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn732(523)A | electrostatic stabiliser |
| Asn732(523)A | enhance reactivity |
| Tyr609(400)A | hydrogen bond donor |
| His689(480)A | proton acceptor |
Chemical Components
proton transfer, hydride transfer, overall product formed, overall reactant used, ingold: bimolecular nucleophilic addition
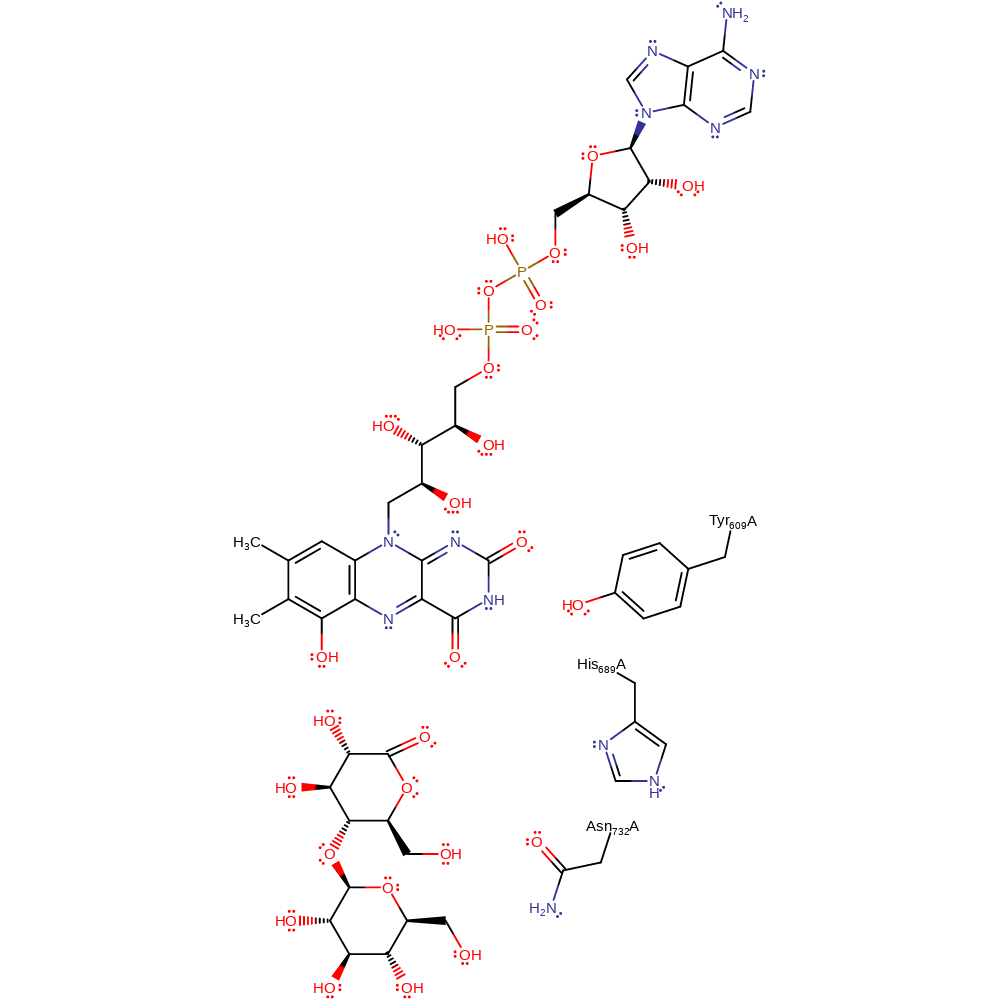
Step 2. Tyr609 interacts with the water molecule near the protonated His689, promoting deprotonation and restoring His689 to its neutral state. Following this step FADH2 is oxidised back to FAD using a haem and oxygen.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr609(400)A | activator, hydrogen bond donor |
| Asn732(523)A | electrostatic stabiliser |
| His689(480)A | proton donor |
Chemical Components
proton transfer, native state of enzyme regeneratedIntroduction
A radical mechanism is compatible with structure of the active site, although radical species have not been detected spectroscopically. The first step involves the transfer of one electron from the 1-hydroxyl oxygen of the substrate to N5 of FAD, along with the abstraction of the hydroxyl proton by His689. The second step involves the transfer of the 1H as a radical to flavin radical intermediate.
Catalytic Residues Roles
| UniProt | PDB* (1kdg) | ||
| His707 | His689(480)A | Acts as a general base to accept a proton from the hydroxyl group of the substrate. | proton acceptor, electrostatic stabiliser, proton donor |
| Tyr627 | Tyr609(400)A | Promotes deprotonation of protonated His689 after the first step of the mechanism. | activator, electrostatic stabiliser |
| Asn750 | Asn732(523)A | Forms a hydrogen bond to the oxygen of the hydroxyl group on the substrate, promoting deprotonation. | activator, electrostatic stabiliser |
Chemical Components
overall reactant used, radical formation, electron transfer, radical termination, overall product formed, bimolecular homolytic elimination, proton transfer, native state of enzyme regeneratedReferences
- Hallberg BM et al. (2003), J Biol Chem, 278, 7160-7166. Mechanism of the Reductive Half-reaction in Cellobiose Dehydrogenase. DOI:10.1074/jbc.m210961200. PMID:12493734.
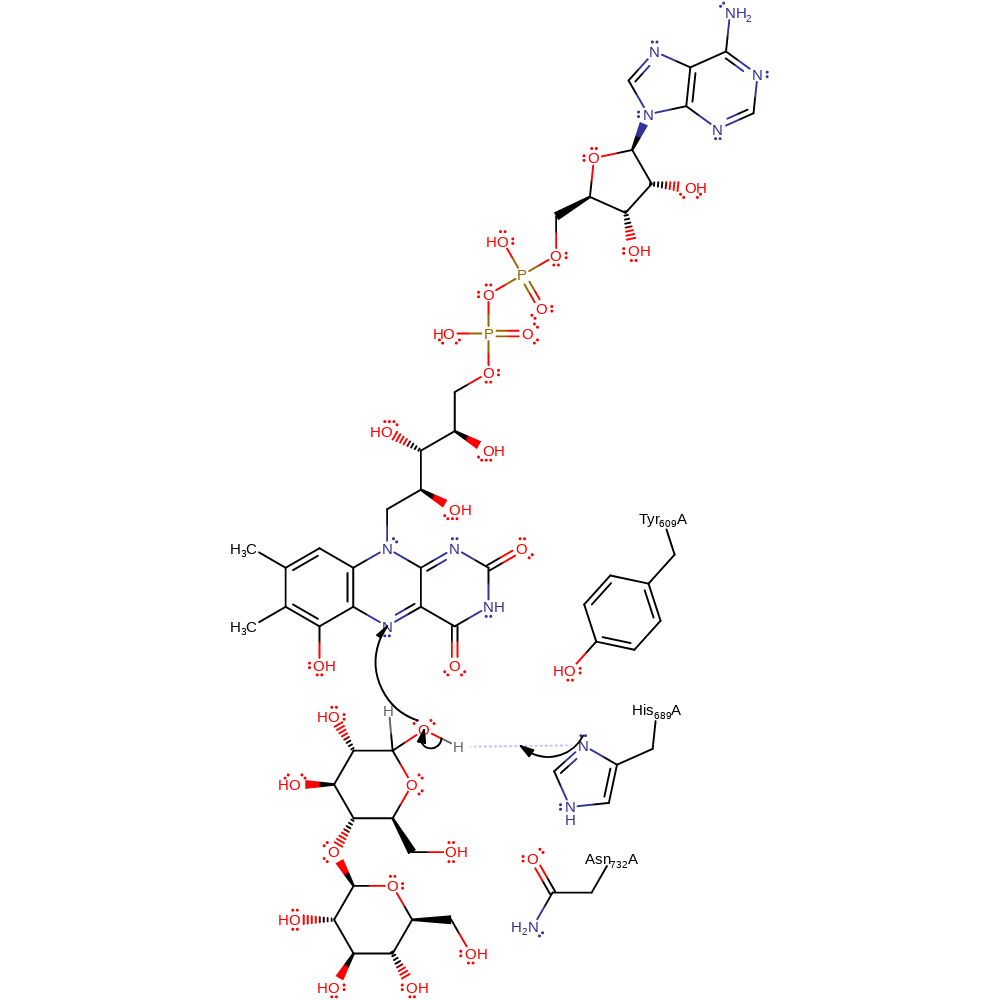
Step 1. An electron from the substrate hydroxyl oxygen may be transferred to the flavin C4a or N5. The hydroxyl proton is abstracted by His689, forming a flavin radical and a substrate radical.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn732(523)A | activator |
| Tyr609(400)A | electrostatic stabiliser |
| His689(480)A | proton acceptor |
Chemical Components
overall reactant used, radical formation, electron transfer
Step 2. A hydrogen radical is transferred from the substrate radical to the flavin radical, forming the product and FADH2.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr609(400)A | electrostatic stabiliser |
| His689(480)A | electrostatic stabiliser |
| Asn732(523)A | electrostatic stabiliser |
Chemical Components
radical termination, overall product formed, ingold: bimolecular homolytic elimination
Step 3. Tyr609 promotes the deprotonation of His689 by a water molecule. FADH2 is oxidised back to FAD by a haem group and oxygen.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr609(400)A | activator |
| His689(480)A | proton donor |




 Download:
Download:  Download:
Download: