N-carbamoyl-D-amino-acid hydrolase
N-carbomyl-D-amino-acid amidohydrolase catalyses the hydrolysis of N-carbamoyl-D-amino acids to the corresponding D-amino acids which are useful intermediates in the preparation of beta-lactam antibiotics. Industrial production of beta-lactam antibiotics is now being developed using this enzyme.
Reference Protein and Structure
- Sequence
-
Q44185
 (3.5.1.77)
(3.5.1.77)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Agrobacterium tumefaciens (Bacteria)

- PDB
-
1fo6
- CRYSTAL STRUCTURE ANALYSIS OF N-CARBAMoYL-D-AMINO-ACID AMIDOHYDROLASE
(1.95 Å)



- Catalytic CATH Domains
-
3.60.110.10
 (see all for 1fo6)
(see all for 1fo6)
Enzyme Reaction (EC:3.5.1.77)
Enzyme Mechanism
Introduction
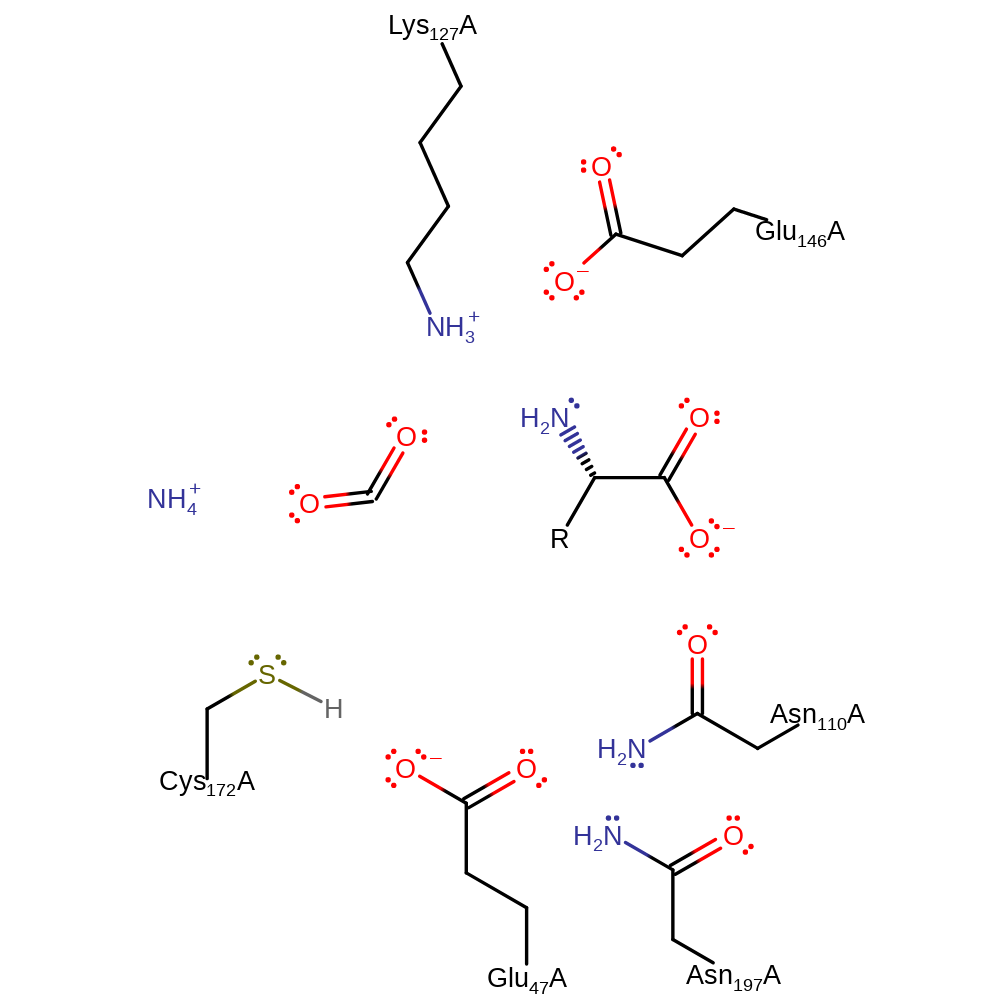
N-carbomyl-D-amino-acid amidohydrolase catalyses the hydrolysis of N-carbamoyl-D-amino acids to the corresponding D-amino acids with the release of carbon dioxide and ammonia. The catalytic mechanism of N-carbomyl-D-amino-acid amidohydrolase begins with an acylation reaction. The sulphur atom of Cys 172 makes a nucleophilic attack on the C atom of the substrate carbonyl group, with Glu 47 acting as a general base in removing the proton from the sulphur atom. Lys 127 can then stabilise the tetrahedral transition state, and cleavage of the C-N bond occurs by the action of Glu 47 as a general acid catalyst to release ammonia. Deacylation of the acyl-enzyme intermediate occurs by Glu 47 acting as a general base to activate a water molecule for nucleophilic attack of the carbonyl carbon. The transition state is again stabilised by Lys 127 and Glu 47 again donates a proton, this time to Cys 172, which releases an acid component that decomposes further to release carbon dioxide leaving the D-amino acid product.
Catalytic Residues Roles
| UniProt | PDB* (1fo6) | ||
| Glu47 | Glu47A | It deprotonates Cys 171 to allow its nucleophilic attack on the substrate carbonyl carbon. It protonates the leaving group. It activates a water molecule to restore the enzyme from the acylenzyme intermediate. | proton acceptor, proton donor |
| Lys127 | Lys127A | It forms an oxyanion hole to stabilise the negatively charged transition state. | electrostatic stabiliser |
| Glu146 | Glu146A | Stabilises Lys 127 through hydrogen bonding. | electrostatic stabiliser |
| Cys172 | Cys172A | It's thiol group acts as a nucleophile to attack the carbonyl carbon of the substrate to form an acylenzyme intermediate. | nucleofuge, nucleophile, proton acceptor, proton donor |
| Asn110, Asn197 | Asn110A, Asn197A | Stabilises Glu 47 through hydrogen bonding which lowers the pKa of Glu 47 so it more willingly accepts a proton. | electrostatic stabiliser |
Chemical Components
proton transfer, bimolecular nucleophilic addition, enzyme-substrate complex formation, intermediate formation, overall reactant used, rate-determining step, unimolecular elimination by the conjugate base, intermediate collapse, enzyme-substrate complex cleavage, native state of enzyme regenerated, heterolysis, inferred reaction step, overall product formedReferences
- Wang WC et al. (2001), J Mol Biol, 306, 251-261. Crystal structure and site-directed mutagenesis studies of N-carbamoyl-d-amino-acid amidohydrolase from Agrobacterium radiobacter reveals a homotetramer and insight into a catalytic cleft11Edited by R. Huber. DOI:10.1006/jmbi.2000.4380. PMID:11237598.
- Han W et al. (2009), Chem Phys Lett, 472, 107-112. The substrate specificity and the catalytic mechanism of N-carbamyl- d -amino acid amidohydrolase: A theoretical investigation. DOI:10.1016/j.cplett.2009.01.086.
- Chen CY et al. (2003), J Biol Chem, 278, 26194-26201. Structural Basis for Catalysis and Substrate Specificity of Agrobacterium radiobacter N-Carbamoyl-D-amino Acid Amidohydrolase. DOI:10.1074/jbc.m302384200. PMID:12709423.
- Grifantini R et al. (1996), J Biol Chem, 271, 9326-9331. Topological Mapping of the Cysteine Residues of N-Carbamyl-D-amino-acid Amidohydrolase and Their Role in Enzymatic Activity. DOI:10.1074/jbc.271.16.9326. PMID:8621596.

Step 1. Glu47 deprotonates Cys172 which activates it nucleophilically attack the carbon of the carbonyl group.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn197A | electrostatic stabiliser |
| Lys127A | electrostatic stabiliser |
| Asn110A | electrostatic stabiliser |
| Glu146A | electrostatic stabiliser |
| Glu47A | proton acceptor |
| Cys172A | nucleophile, proton donor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, enzyme-substrate complex formation, intermediate formation, overall reactant used, rate-determining step
Step 2. The oxyanion initiates an elimination which results in the cleavage of the amino group which is protonated by Glu47 to produce ammonia.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn110A | electrostatic stabiliser |
| Lys127A | electrostatic stabiliser |
| Glu146A | electrostatic stabiliser |
| Asn197A | electrostatic stabiliser |
| Glu47A | proton donor |
Chemical Components
ingold: unimolecular elimination by the conjugate base, proton transfer, intermediate collapse
Step 3. Glu47 deprotonates a water molecule which activates it to nucleophilically attack the carbon of the carbonyl group.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn110A | electrostatic stabiliser |
| Lys127A | electrostatic stabiliser |
| Glu146A | electrostatic stabiliser |
| Asn197A | electrostatic stabiliser |
| Glu47A | proton acceptor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, intermediate formation, overall reactant used
Step 4. The oxyanion once again initiates an elimination which results in the cleavage of the acyl-enzyme and Cys172 is protonated by Glu47 which returns the active site to its native state.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn110A | electrostatic stabiliser |
| Lys127A | electrostatic stabiliser |
| Glu146A | electrostatic stabiliser |
| Asn197A | electrostatic stabiliser |
| Cys172A | nucleofuge |
| Glu47A | proton donor |
| Cys172A | proton acceptor |
Chemical Components
proton transfer, ingold: unimolecular elimination by the conjugate base, enzyme-substrate complex cleavage, intermediate collapse, intermediate formation, native state of enzyme regenerated
Step 5. The released intermediate further decomposes to release CO2 and the D-amino acid product, which is protonated by the surrounding solvent.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn110A | electrostatic stabiliser |
| Lys127A | electrostatic stabiliser |
| Glu146A | electrostatic stabiliser |
| Asn197A | electrostatic stabiliser |






 Download:
Download: